Basic usage
Linear genome view
To open a linear genome view (LGV), use the menu bar: Add ->
Linear genome view
Scrolling
You can scroll side to side using your mouse wheel or via click and drag. Left and right pan buttons found in the header of the LGV can also be used to scroll in their respective directions.
Zooming
The zoom buttons and the slider bar found in the header of the linear genome view can be used to zoom in and out on the view
You can also
- hold the
Ctrlkey and use your mousewheel or trackpad to scroll to zoom in and out of the linear genome view - hold the
Shiftkey and click and drag the linear genome view to create a "rubberband" selection - hold the
Shiftkey without click and drag to just reveal a red vertical guide bar on the linear genome view
Re-ordering tracks
Click and drag up or down on the drag handle on the track labels (indicated by six vertical dots) to reorder tracks.
Re-ordering views
Re-ordering views doesn't use a drag and drop like re-ordering tracks does, but instead, you can click the view menu (the "hamburger" menu that is available for each view) and select "Move up"/"Move down"
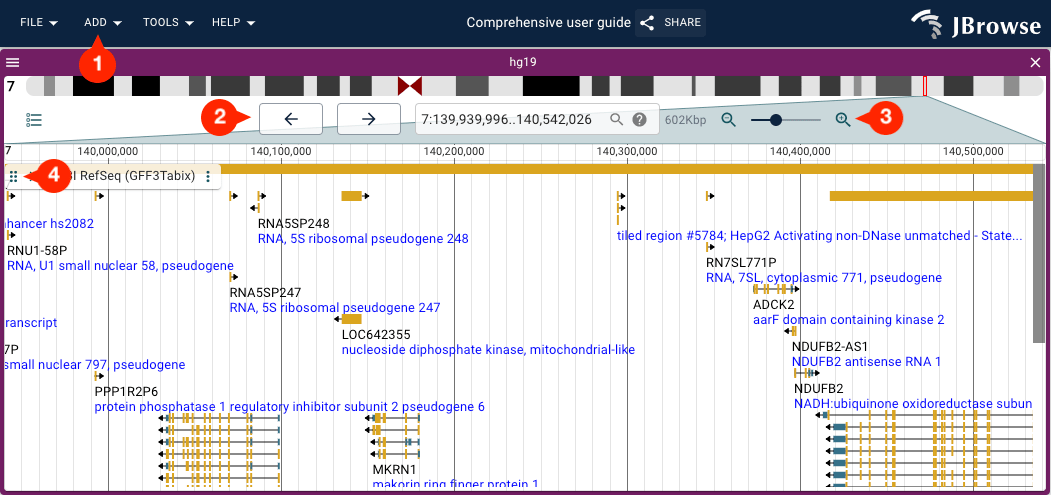
Using the location search box
The location search box is located at the top of the LGV.
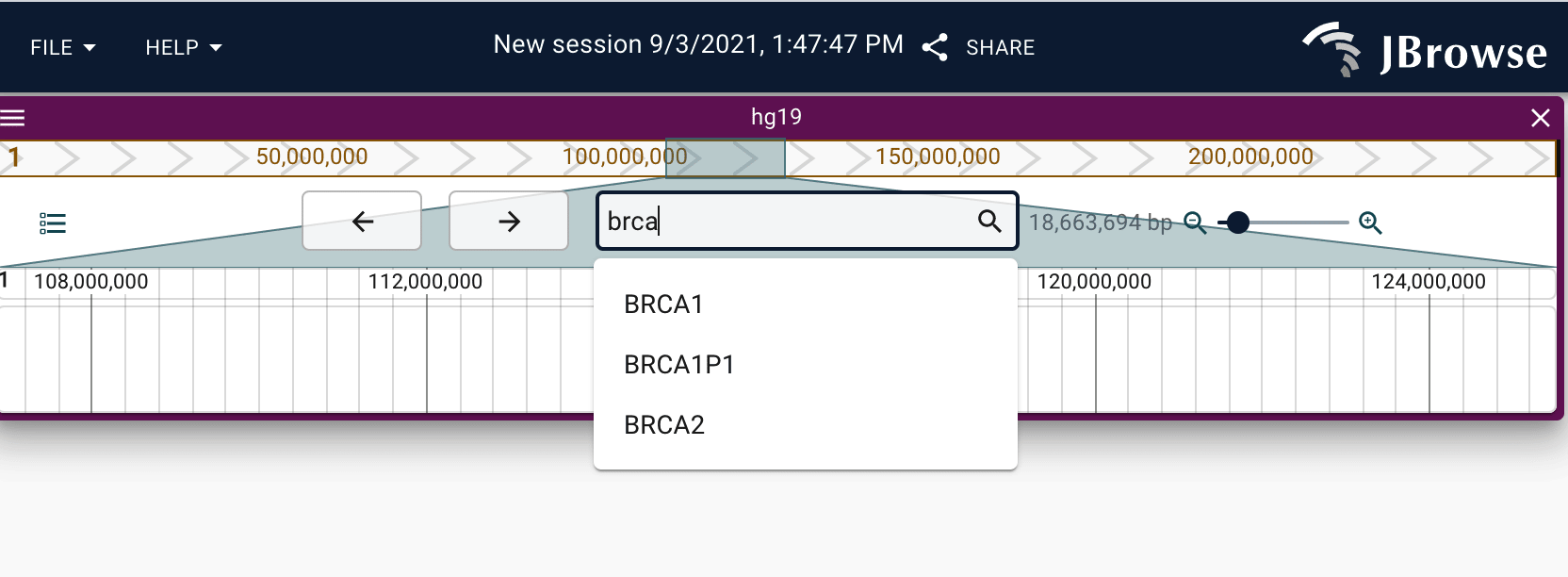
You can search a location in several ways when typing in the search box:
- Searching by region and location, e.g.
chr1:1..100orchr1:1-100orchr1 1 100 - Searching by assembly, region, and location, e.g.
{hg19}chr1:1-100 - Searching discontinuous regions, delimited by a space, and opening them
side-by-side, e.g.
chr1:1..100 chr2:1..100 - Searching in any of the above ways and appending [rev] to the end of the
region will horizontally flip it, e.g.
chr1:1-100\[rev\] - If configured, searching by gene name or feature keywords, e.g.
BRCA1
To configure name searching, you or the admin of the instance will need to create a "text index". See the configuration guide for more information.
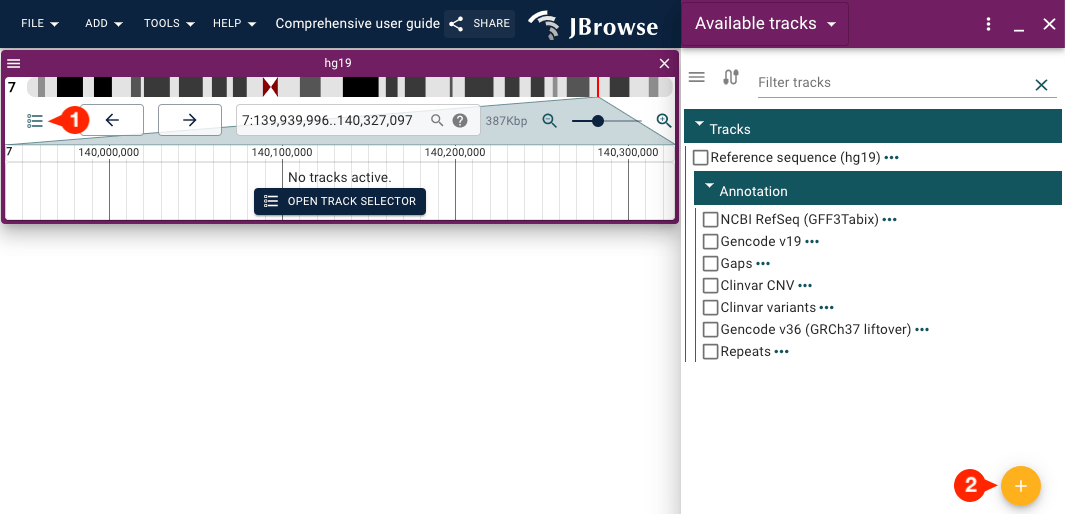
Opening tracks
To open a new track or connection, use the menu bar: File -> Open track..
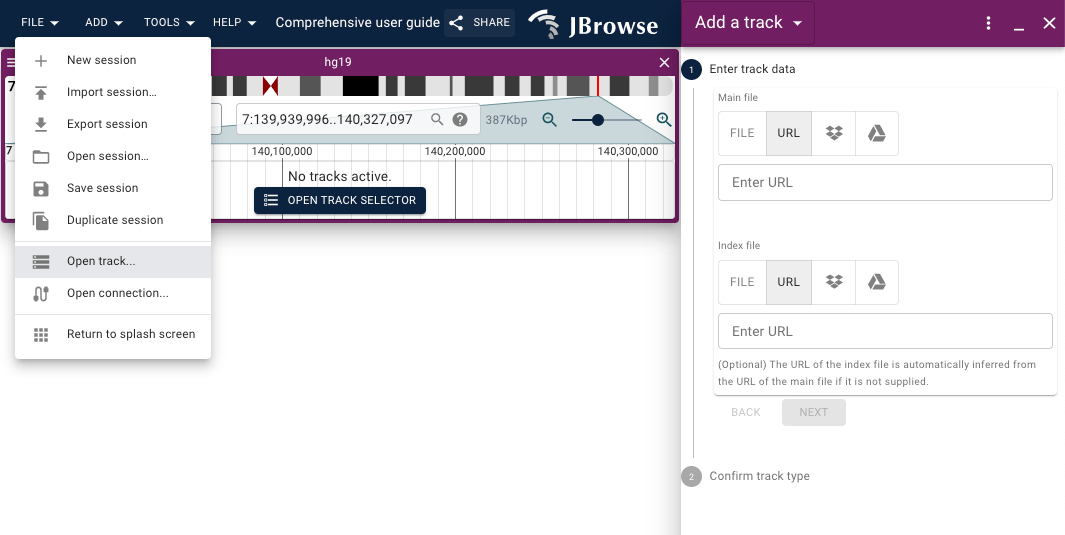
There is a circular plus (+) icon button inside the "Available tracks" widget that can also be used to access the "Add a track" form.
In the "Add a track" form, you can provide a URL to a file to load, or you can also open files from your local machine. In some cases, you need to provide an index (bigwig files for example have no index, but BAM/CRAM or tabix filetypes like VCF/GFF/BED tabix do). In some cases we can automatically infer the index e.g. if you provide a URL for a BAM and the index filename is bamfilename +'.bai' but you may need to manually supply it in some cases (index inference can't be done with files from your local machine)
File format support
The following file formats are supported in core JBrowse 2:
General
- CRAM
- BAM
- htsget (requires hand-edited config)
- VCF (Tabix-indexed)
- GFF3 (Tabix-indexed)
- BED (Tabix-indexed)
- BigBed
- BigWig
- BEDPE
- JBrowse 1 nested containment lists (NCList)
- .hic (Hi-C contact matrix visualization)
SV inspector
- plain text VCF, BED, CSV, TSV, BEDPE, STAR-fusion output (tabular formats)
Synteny and dotplot
- PAF (e.g. minimap2)
- .delta (MUMmer)
- .anchors (MCScan python version)
- .out (MashMap)
Sequence adapters
- Indexed FASTA
- BGZip indexed FASTA
- 2bit
Additional data formats can be supported via plugins; checkout the plugin store.
For tabix files, TBI or CSI indexes are allowed. CSI or BAI is allowed for BAM. Only CRAI is allowed for CRAM. The index will be inferred for BAI or TBI files as filename+'.bai' for example, but if it is different than this, make sure to specify the index file explicitly.
If you are an administrator, you can add tracks with the command line or with the admin server.
Undo and redo
You can undo the closing of a view, track, or any other action in the UI with the Tools->Undo/Redo buttons. The keyboard shortcut "ctrl+z"/"cmd+z"(mac) work for undo as well as "ctrl+y"/"cmd+shift"z"(mac)
Sharing sessions
On JBrowse Web, the main menu bar has a "Share" button to enable users to share their sessions with other people. The share button generates a URL that can be sent to other users.
You cannot copy the URL in your address bar and send it to other users, you must use the "Share" button to share your session.
Sharing sessions is not available for JBrowse Desktop.
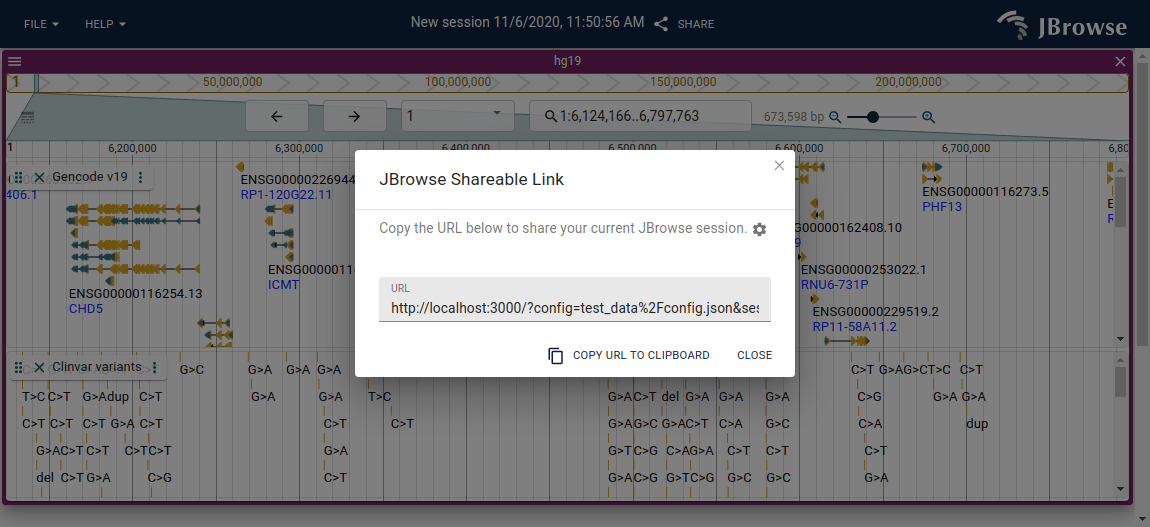
The session URL will contain the following:
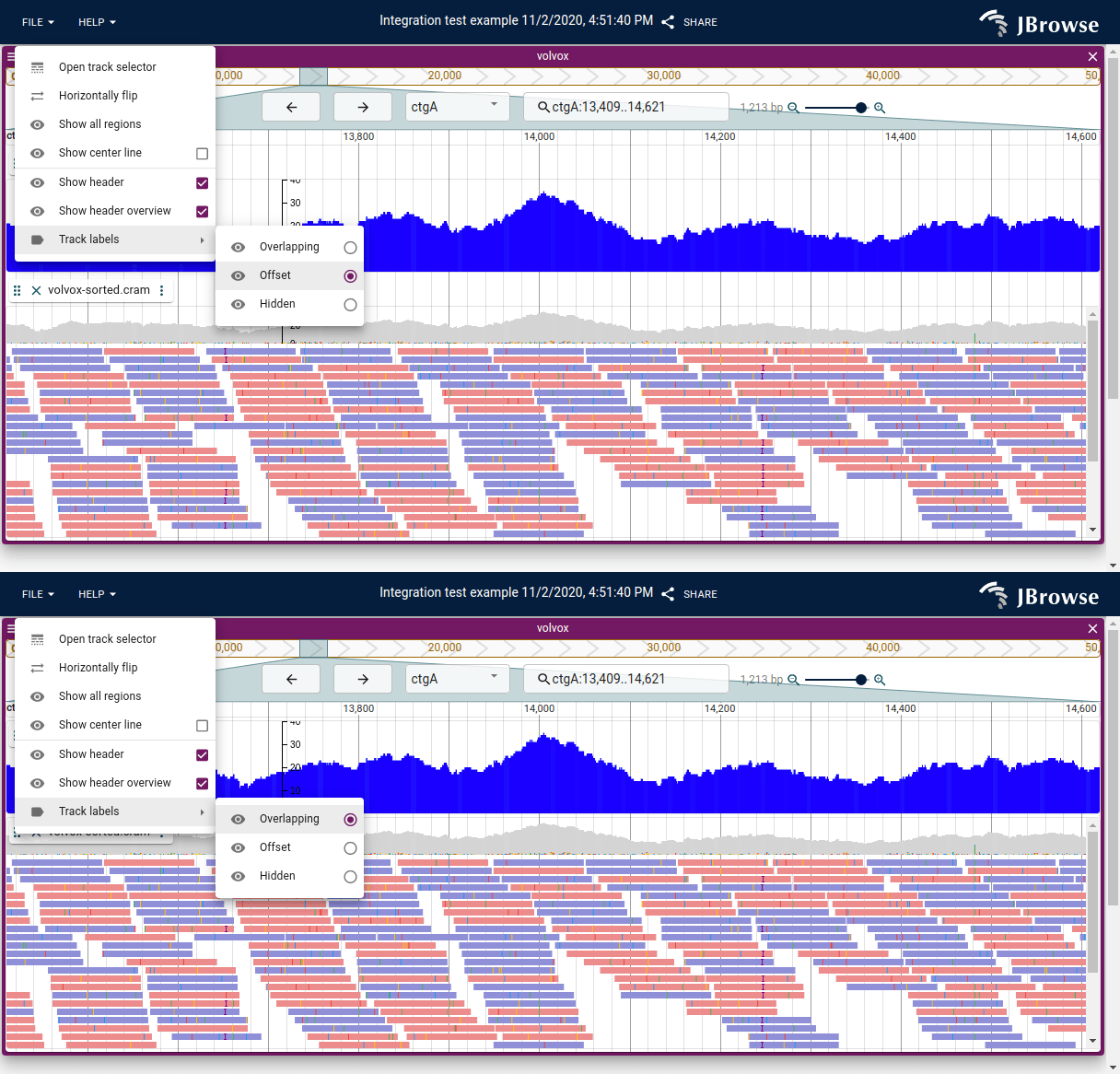
- what views are on the screen, and settings for the views (e.g. track labels overlapping or offset)
- what tracks are in the view
- extra tracks that you added with the "Add track workflow"
- for the alignments track, the show soft clipping and sort settings on the pileup
- ...and more!
This means you can share links with your custom tracks with other users, without being a JBrowse admin!
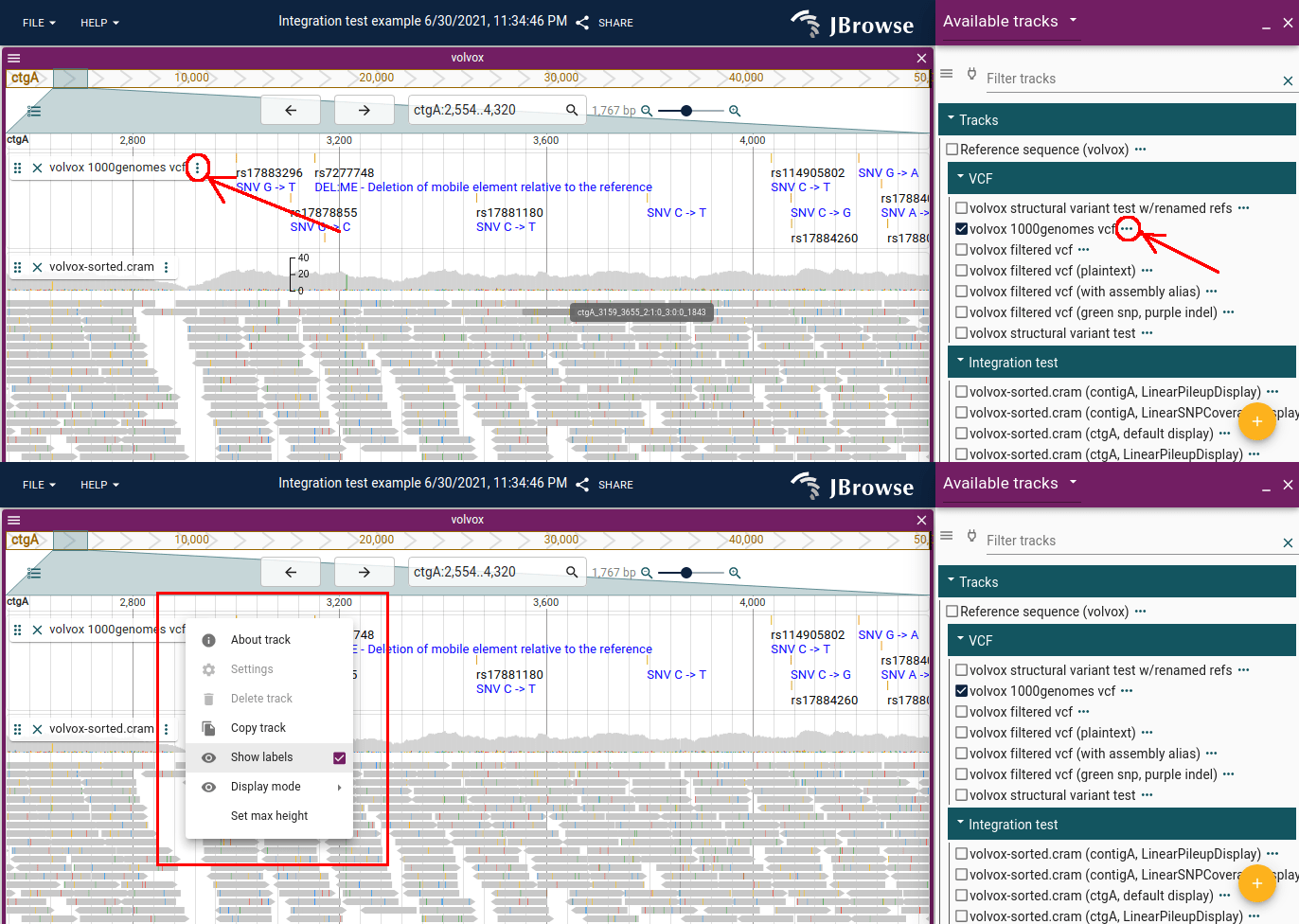
Track menu
Users can access track-specific functions by using the track menu, which is accessible from the track selecter itself ("..." icon) or on the track label (vertical "..."). Some functions are only available when the track is open e.g. from the track label, but more basic options like "About track" are available from the track menu on the track selector.
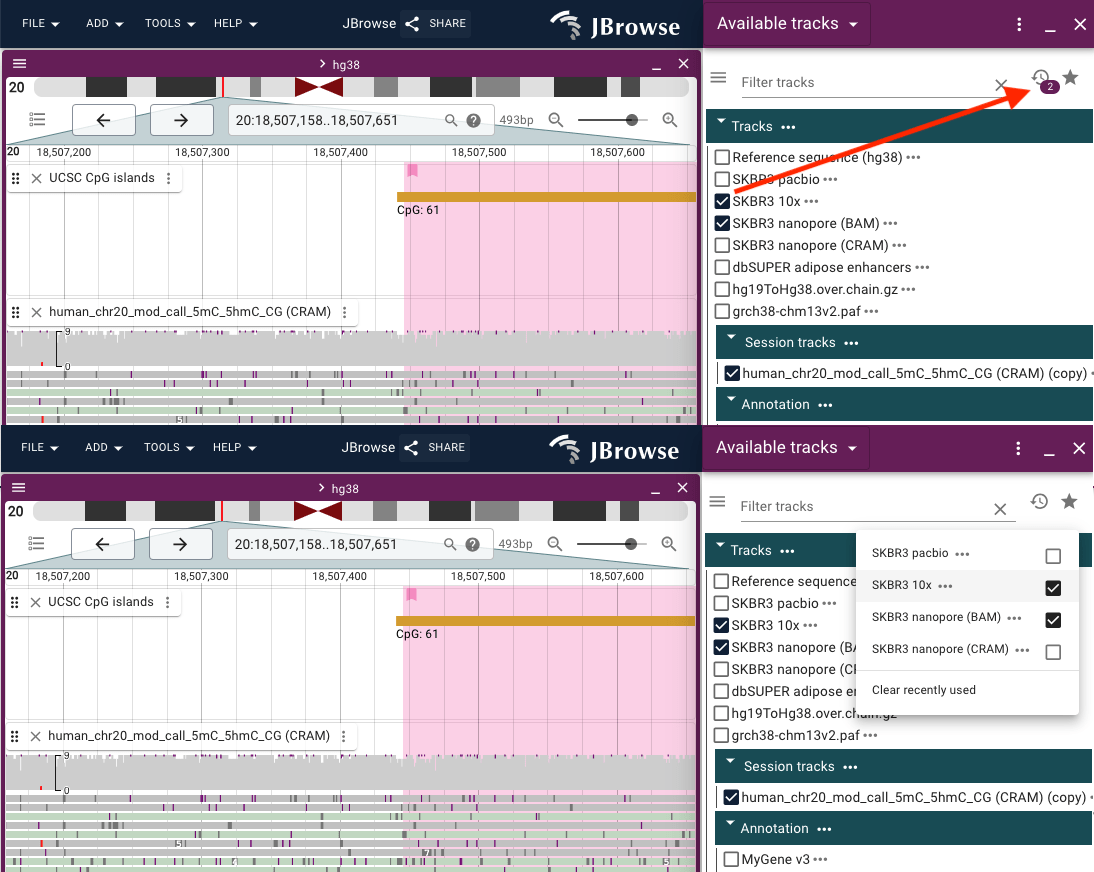
Recently used and Favorite tracks
Users can specify favorite tracks using the Track menu, and view them using the star icon button in the top right corner of the Available tracks widget.
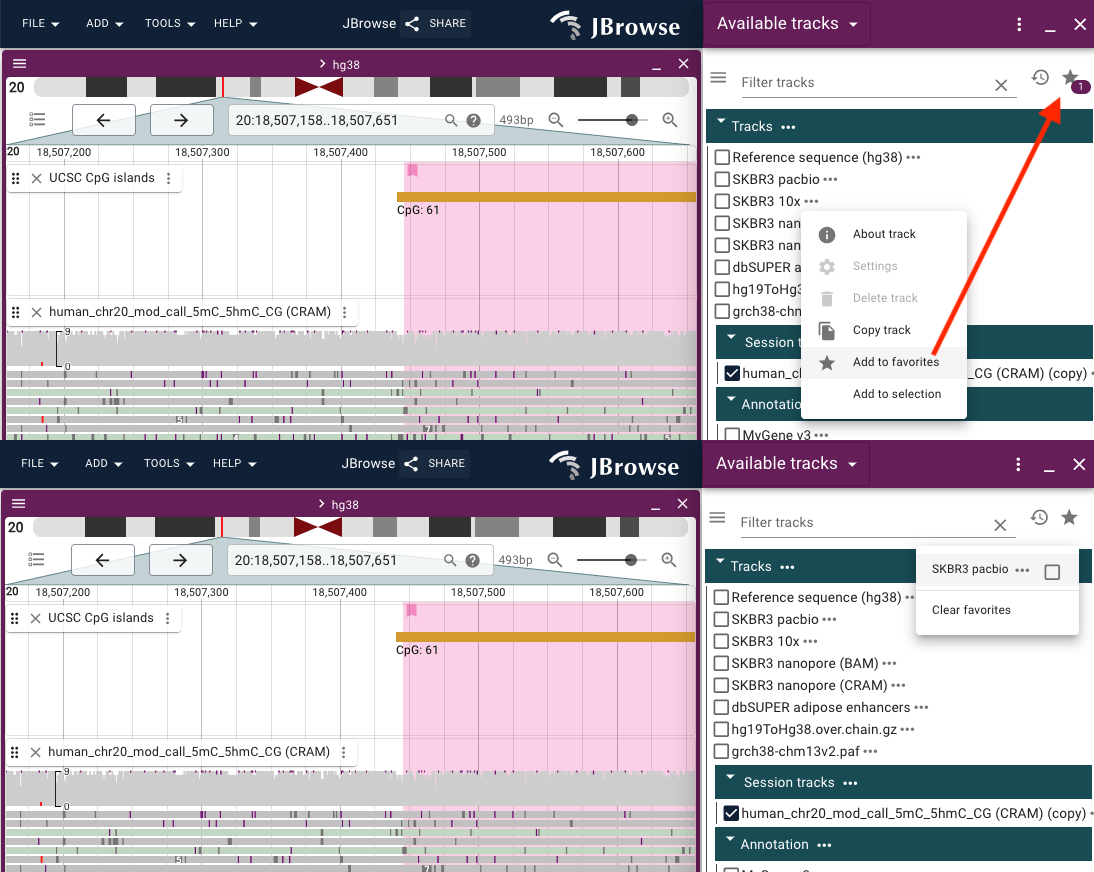
Tracks that have been recently opened will be automatically added to the list of recently used tracks, and can be viewed using the clock icon button in the top right corner of the Available tracks widget.
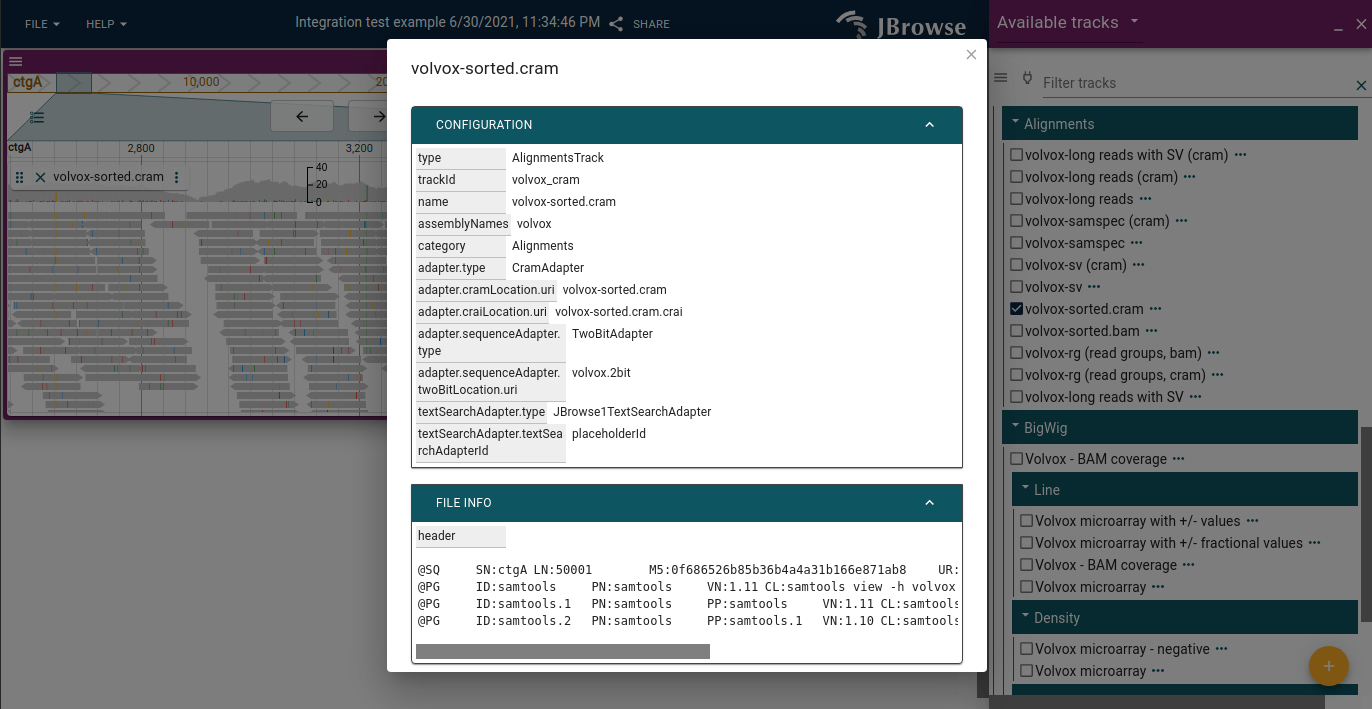
About track dialog
Using the track menu as described above, you can access the "About track" dialog.
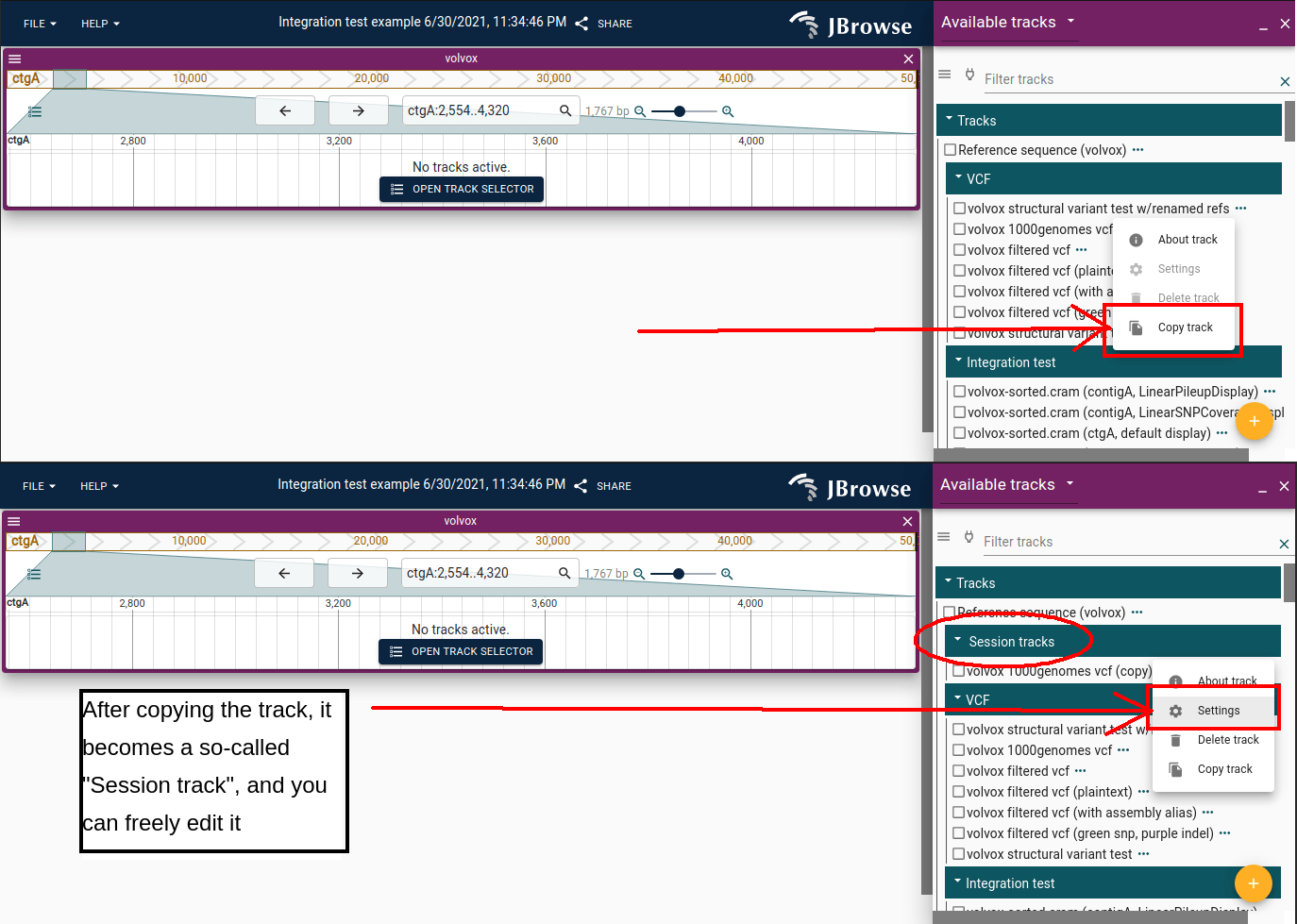
Editing track configs
As a non-admin user, in order to edit a track config, you have to make a copy of the track. This will copy it to your "session tracks", which you can edit freely.
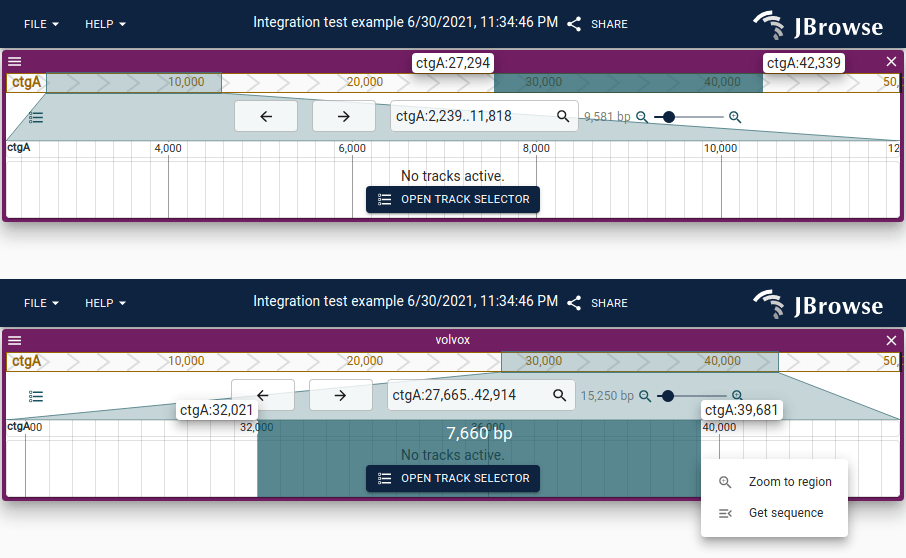
Rubberband selection
The scale bars accept a click-and-drag action to select a region. Rubberband selection can be performed on both the main (lower) and overview (upper) scale bars.
Track label positioning
Track labels can be positioned on their own row or overlapping the data to save vertical screen space. They can also be hidden. This is done by clicking on the hamburger menu for a specific view.
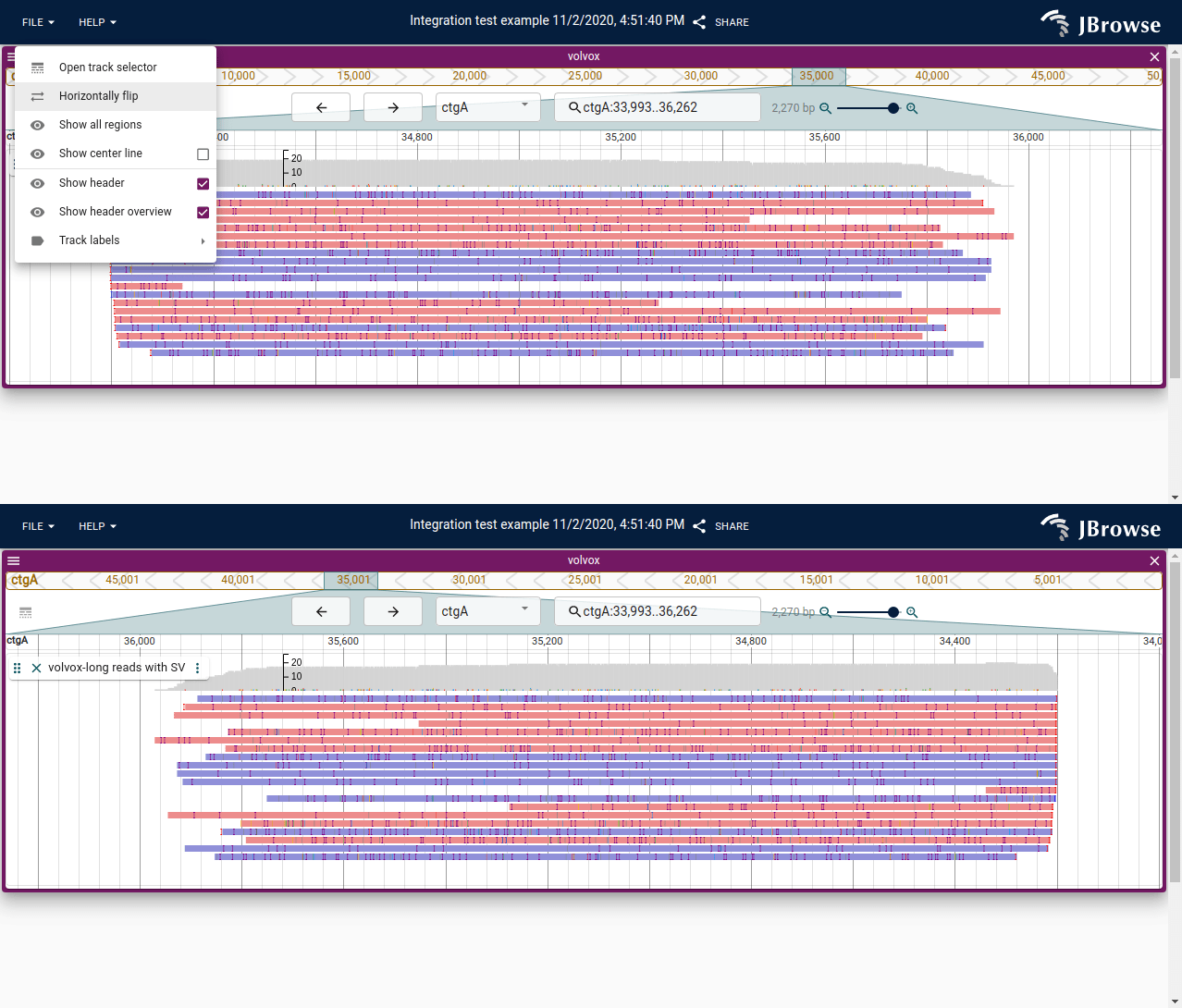
Horizontally flip
The view can be horizontally flipped, or reverse complemented, to make the coordinates go from right to left instead of left to right.
We use triangles pointing in the direction of the orientation in the overview bar to help indicate whether the app is horizontally flipped or not.
Here is an example of before and after horizontally flipping the view:
Toggle drawer widget on left or right side of screen
Using a drop-down menu in the header bar, you can toggle the drawer widget to the left or right side of the screen. It is on the right side by default