JBrowse 2 visualizations gallery
Below are some examples of the visualizations JBrowse 2 has to offer. For live demos, have a look at the demos page.
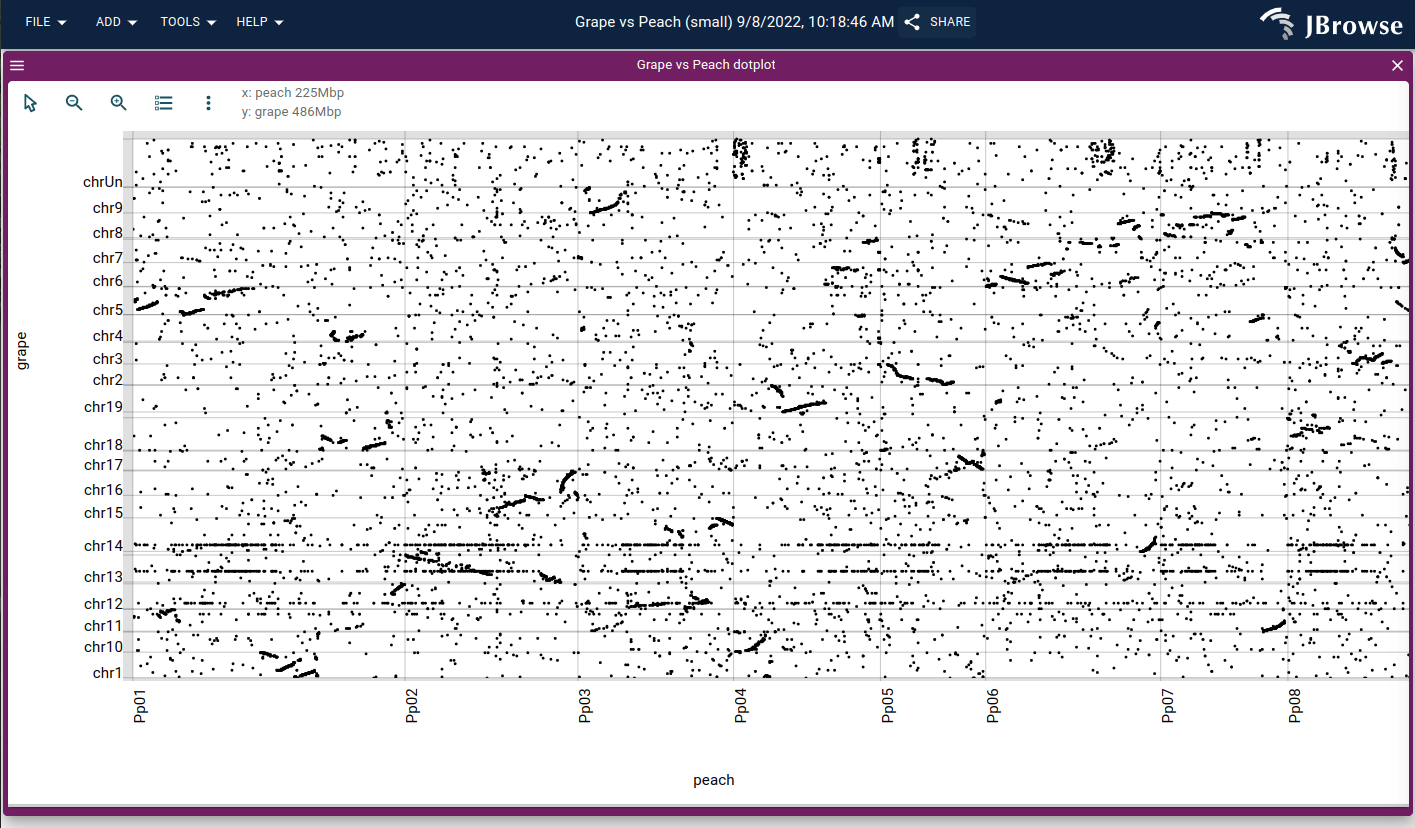
A genome vs genome dotplot displayed in the DotplotView. This displays grape vs peach genomes via PAF file
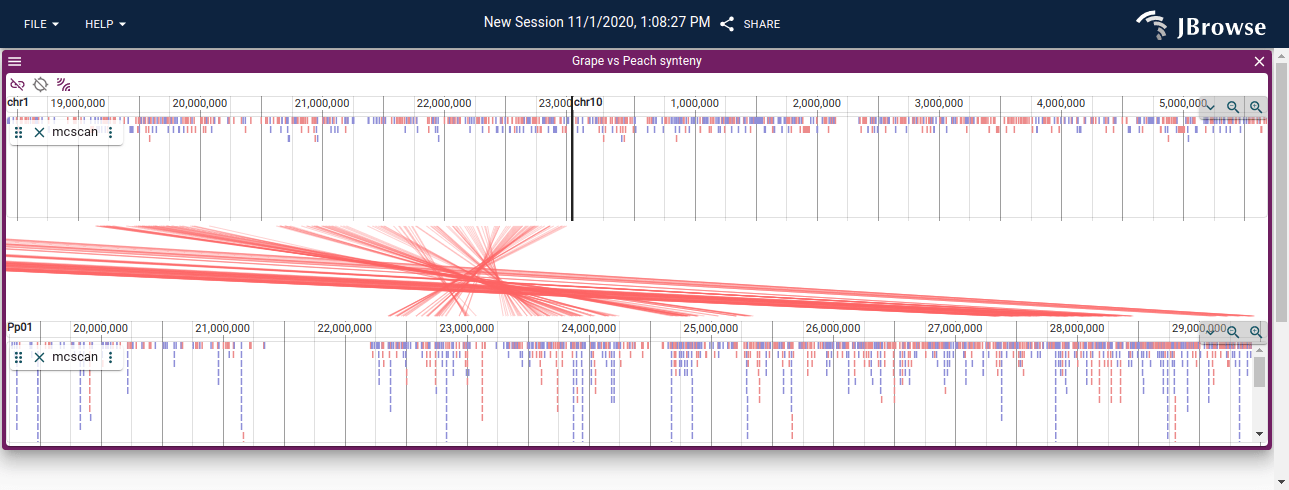
A linear synteny display for grape vs peach displayed in the LinearSyntenyView via MCScan anchors file results
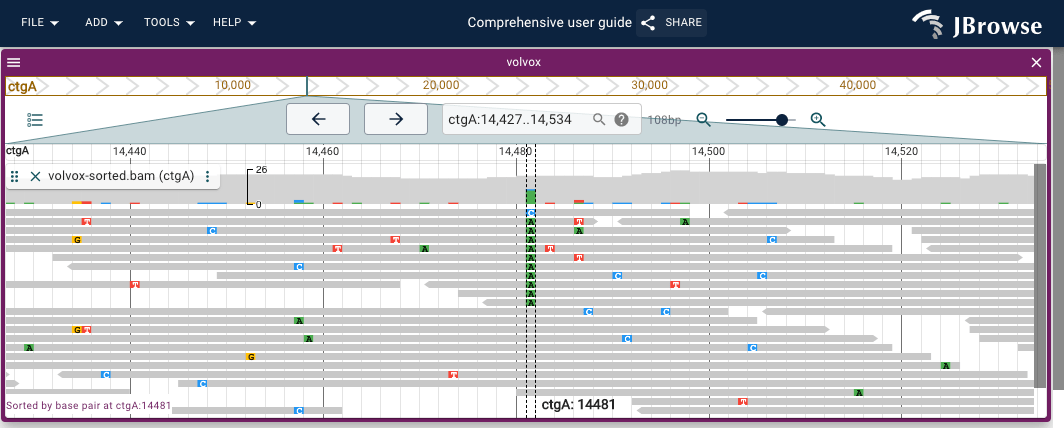
Shows sorting the alignments pileup by the base-pair-at-the-given-position in the AlignmentsTrack
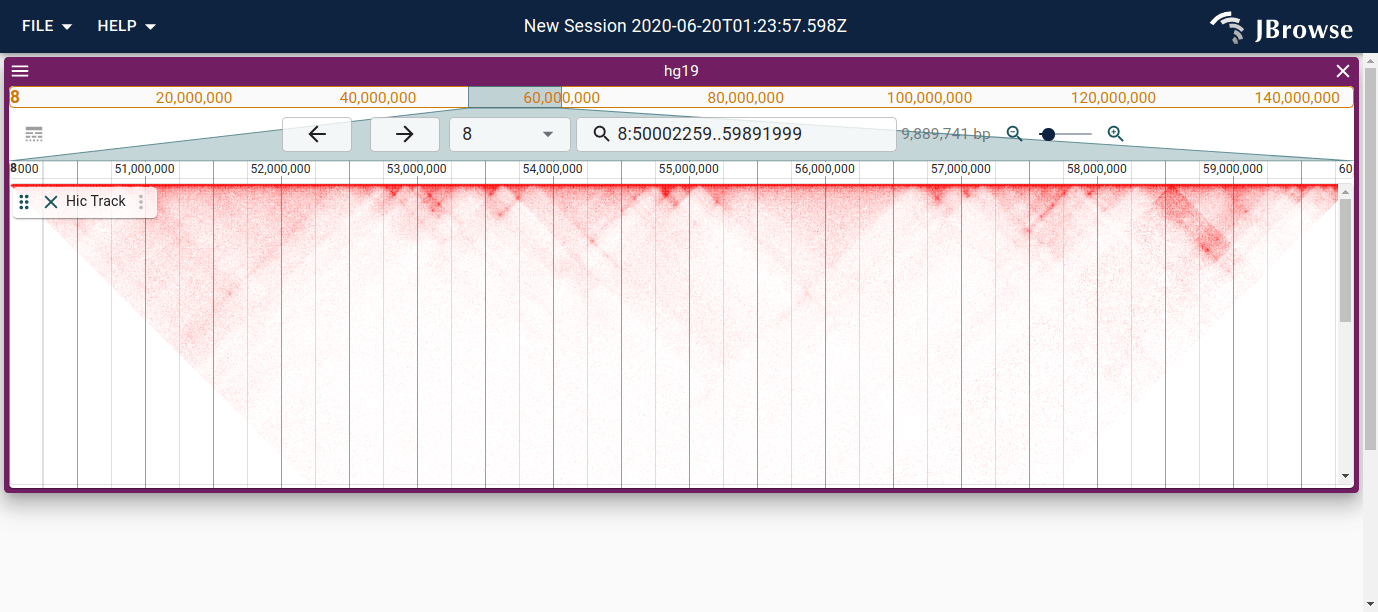
Shows rendering of a .hic file type in the HicTrack
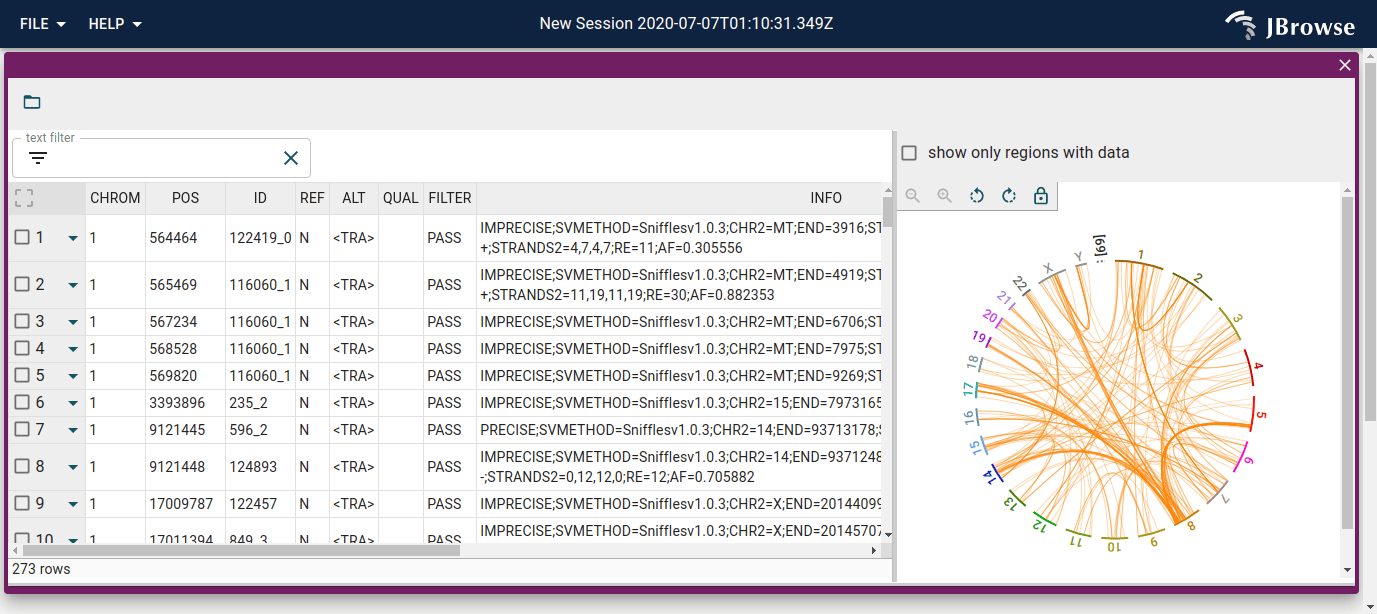
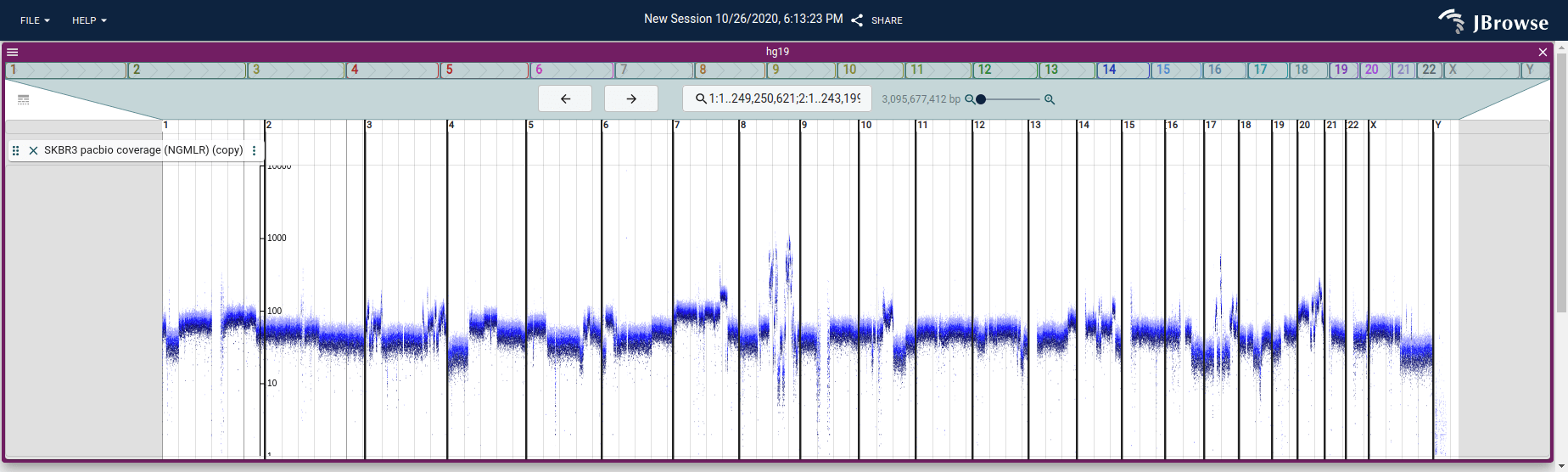
Screenshot of the SV inspector displaying inter-chromosomal translocations in the SKBR3 cell line. The SV inspector is a tabular view plus whole-genome circular overview of the SVs
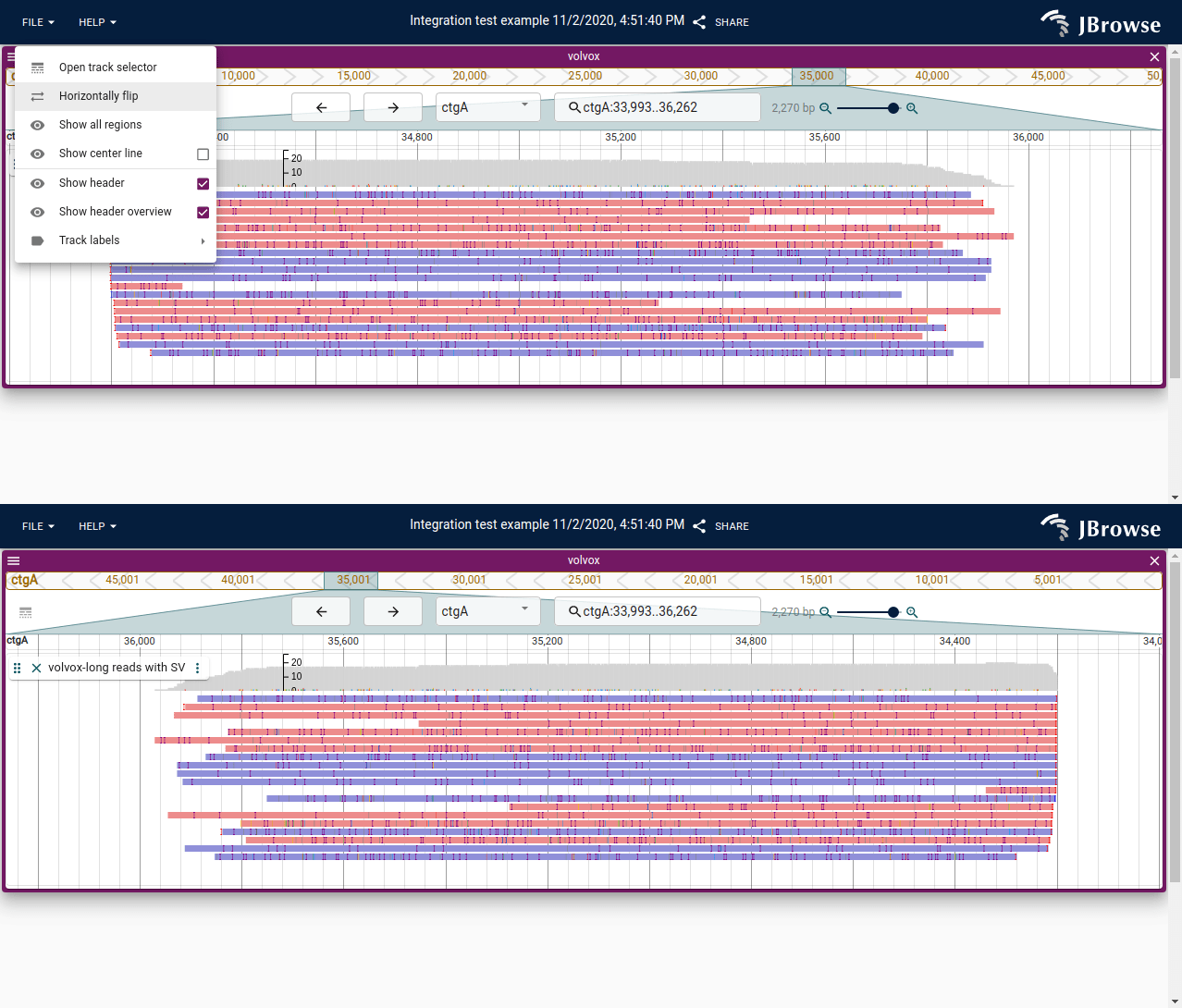
Shows the ability to horizontally flip the current view. This allows the anti-sense strand to be read left to right
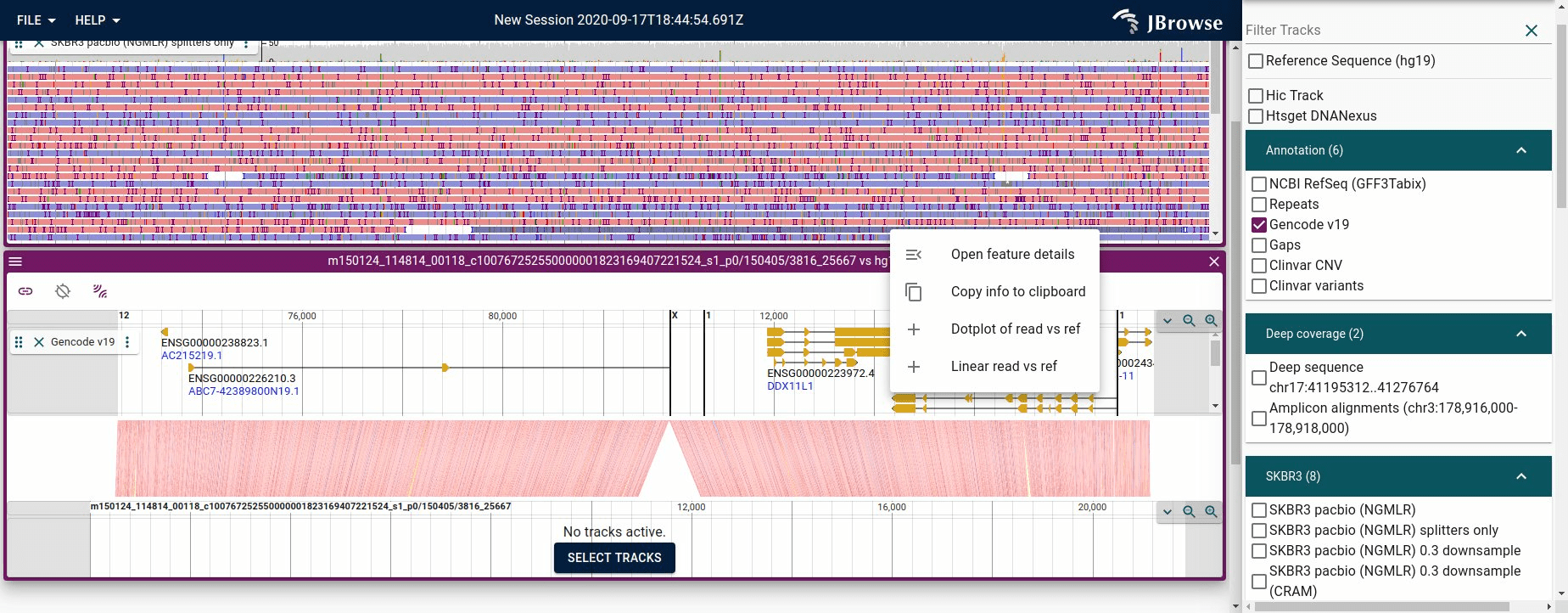
Using the synteny view to render a long-read vs the reference genome. This is available simply by clicking an alignments feature and selecting "Linear read vs ref"
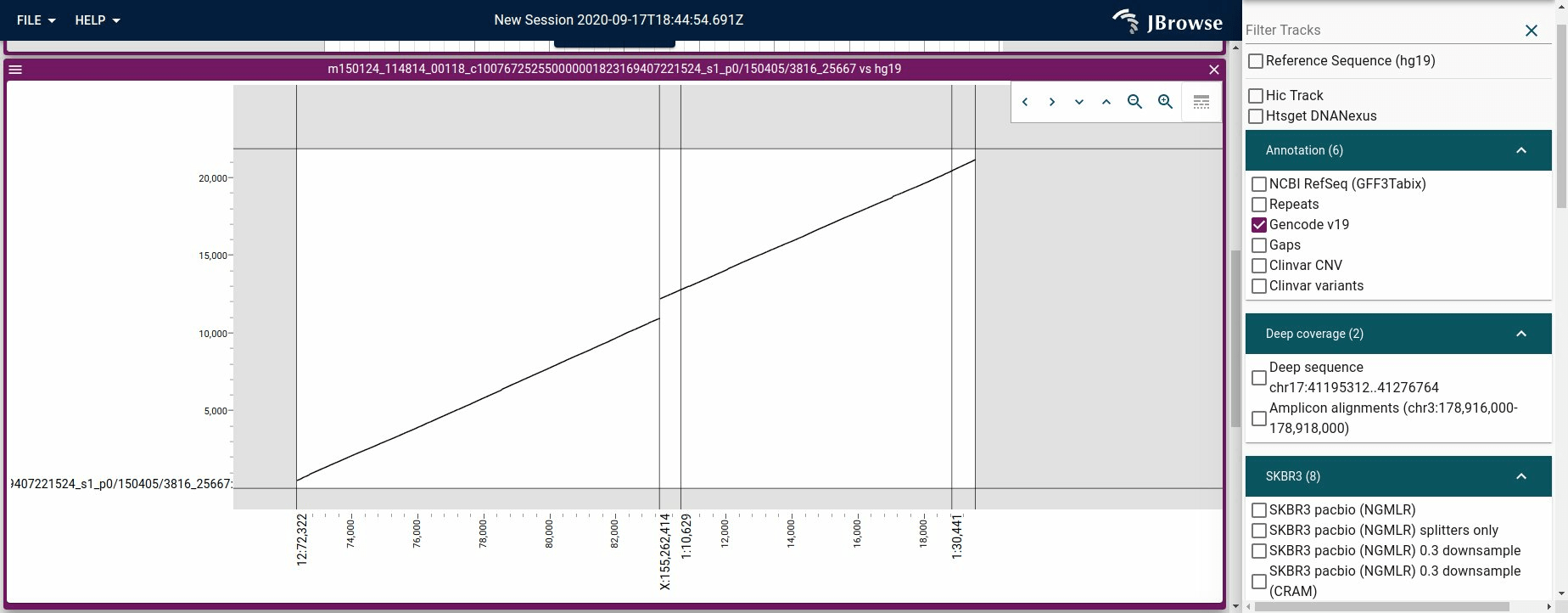
Using the dotplot view to render a long-read vs the reference genome. This is available by right-clicking an alignments feature and selecting "Dotplot read vs ref"
This is a whole-genome overview of CNV data using a BigWig file of read-coverage depth. This uses the XYPlot, with fill turned off. The BigWig data is binned at this resolution, and the blue shades represent the min, mean, and max value in each bin

Image of the "breakpoint split view" which examines the breakpoints of a structural variant, e.g. an interchromosomal translocation, and connects supporting reads (black splines) and the variant call itself (green thicker line, with feet indicating directionality)
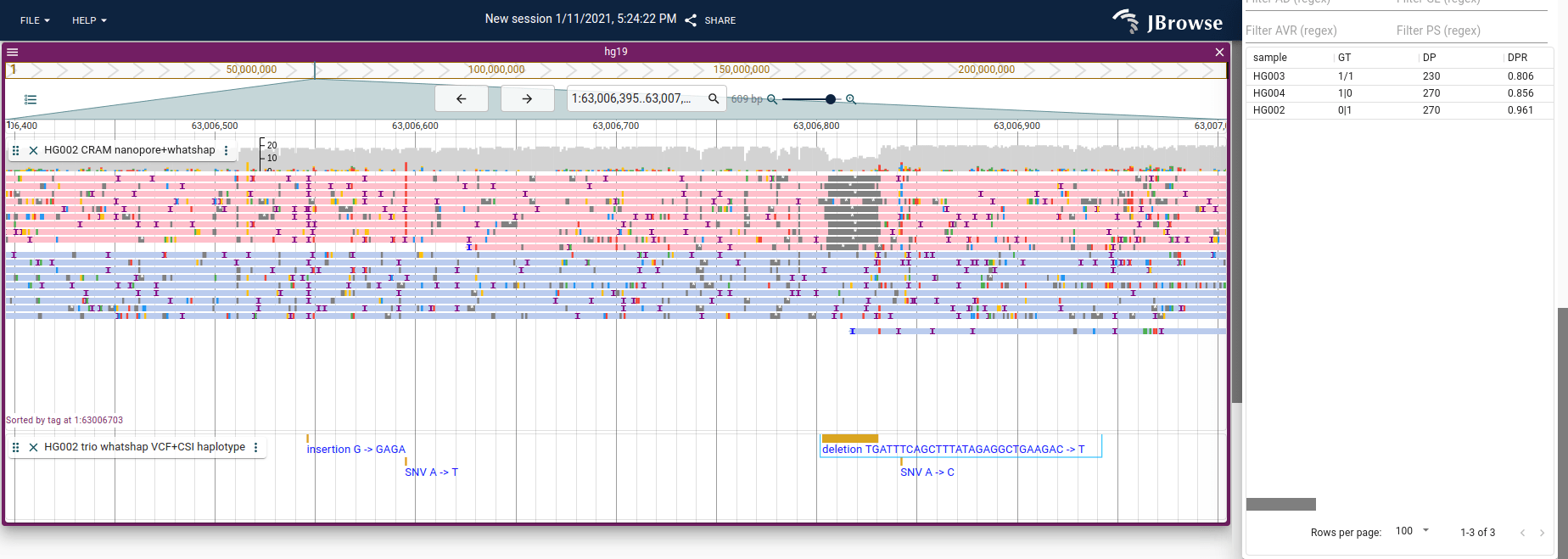
Image of coloring and sorting by the HP (haplotype) tag, which can be done for BAM and CRAM. Users can color, sort, and filter by tags
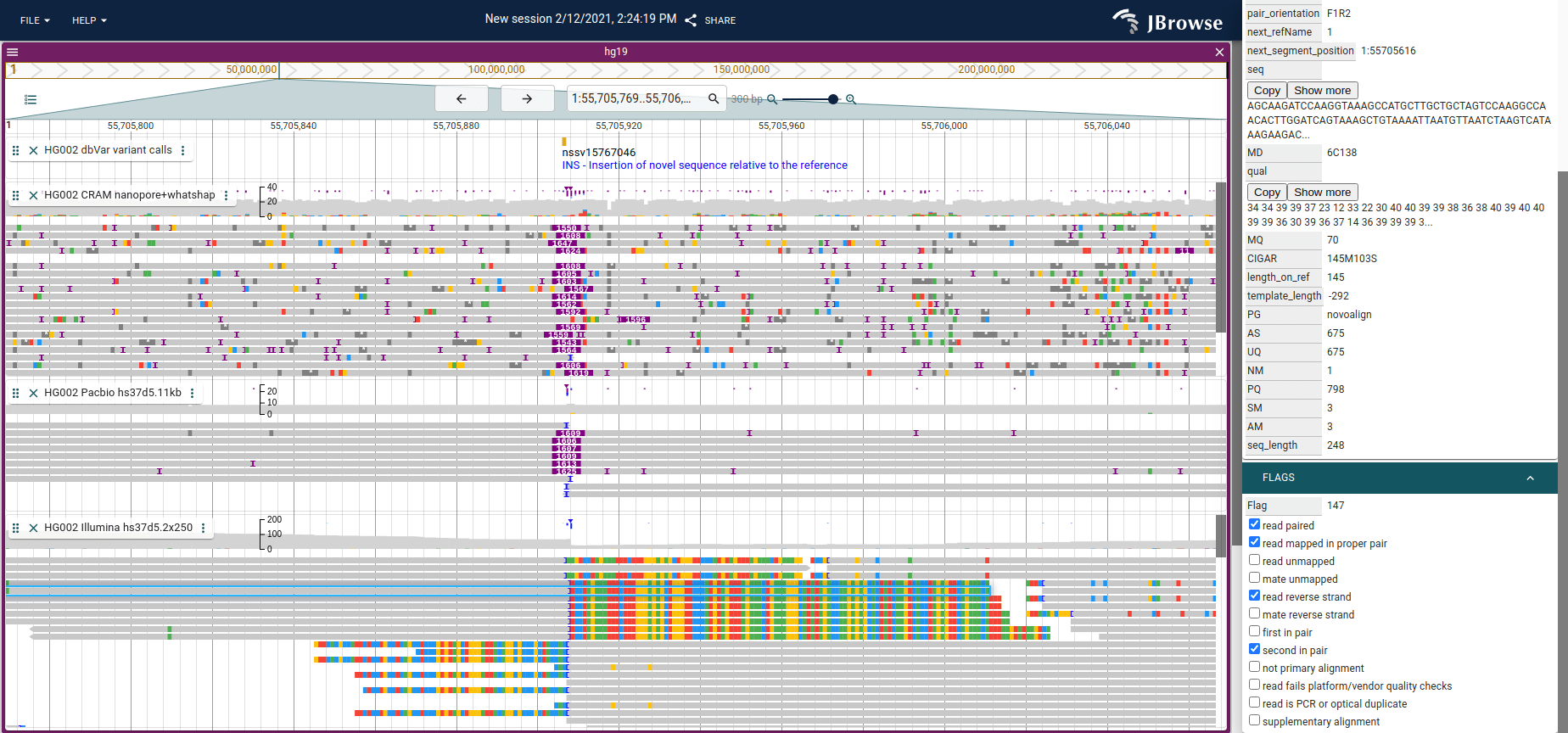
The browser has a variety of ways to help show insertions including "large insertion indicators" (a feature also seen in IGV) and upside down histogram of insertion/clipping counts
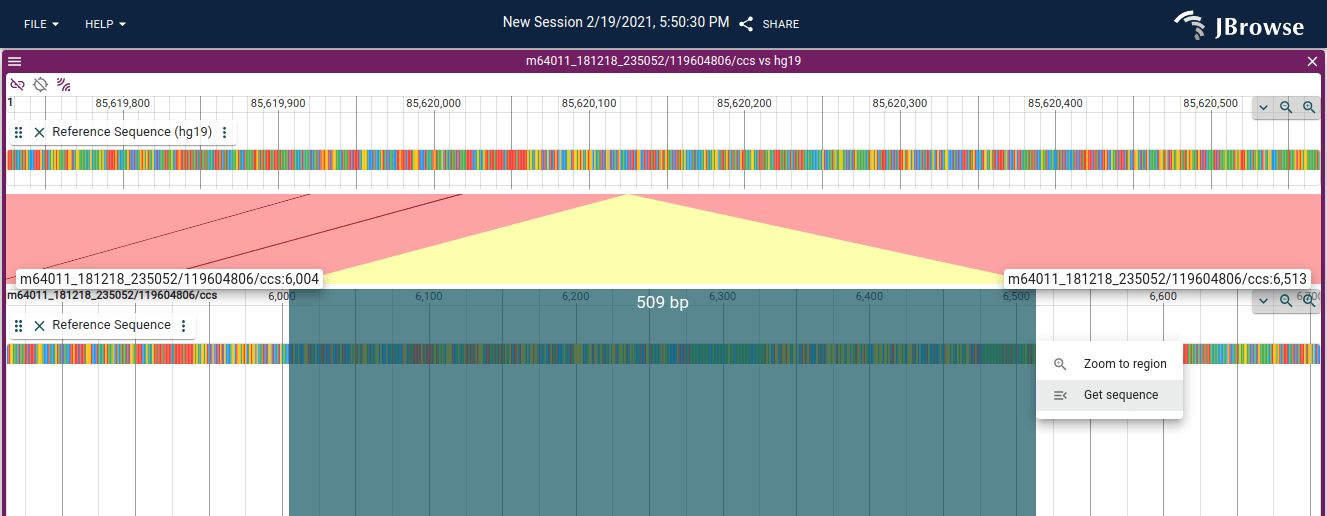
The read vs reference visualization can show the "sequence track" on the "read" to see which bases are inserted. Users can also use the "Get sequence" click and drag to get the sequence of the contents of the insertions or unaligned portions of the read (e.g. softclipped parts)
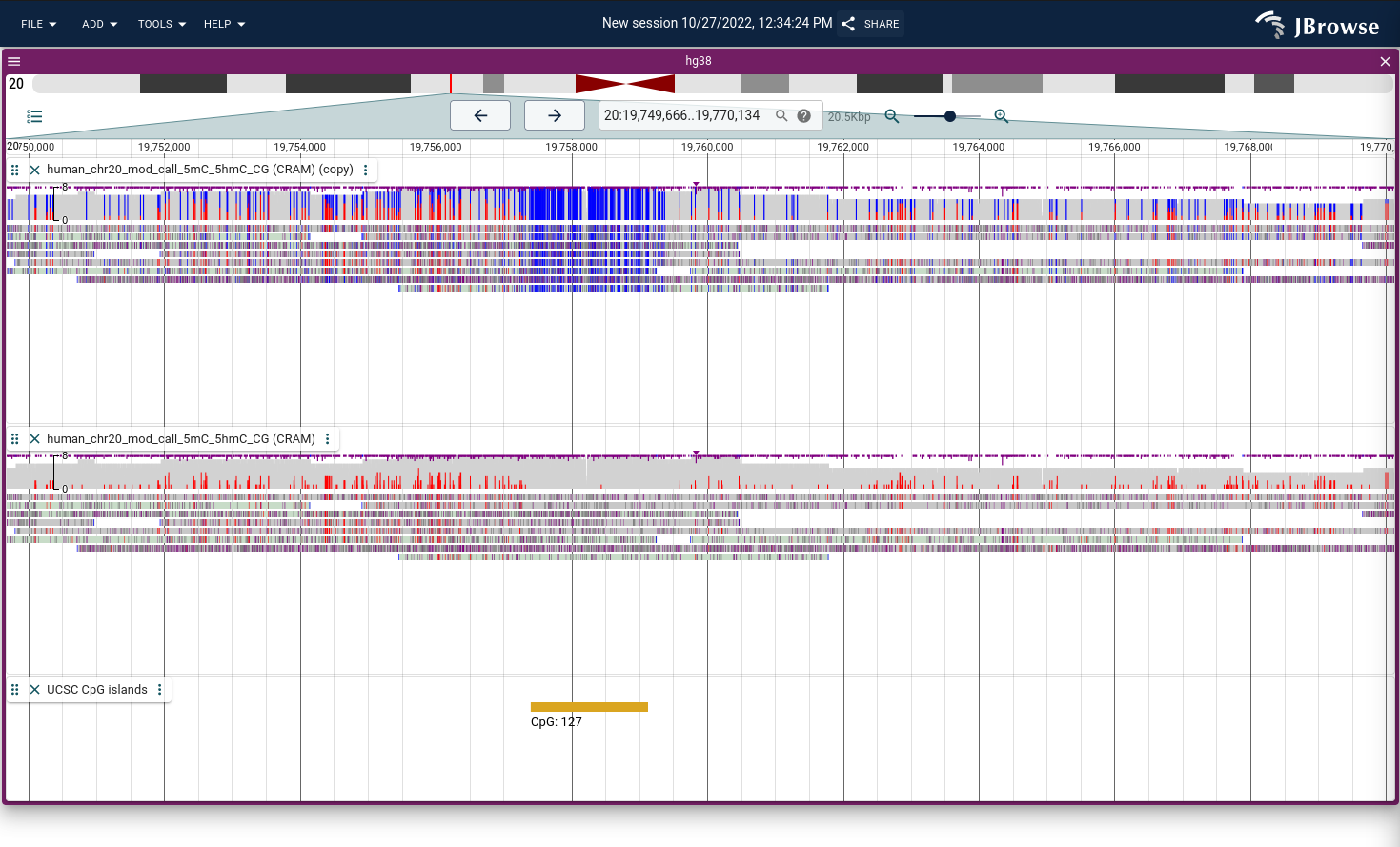
A hypo-methylated CpG island shown using the MM tag with nanopore reads