JBrowse
The next-generation genome browser
JBrowse is a new kind of genome browser that runs on the web, on your desktop, or embedded in your app.
Also check out our latest release blogpost, our embedded components, and our command line tools.
Features
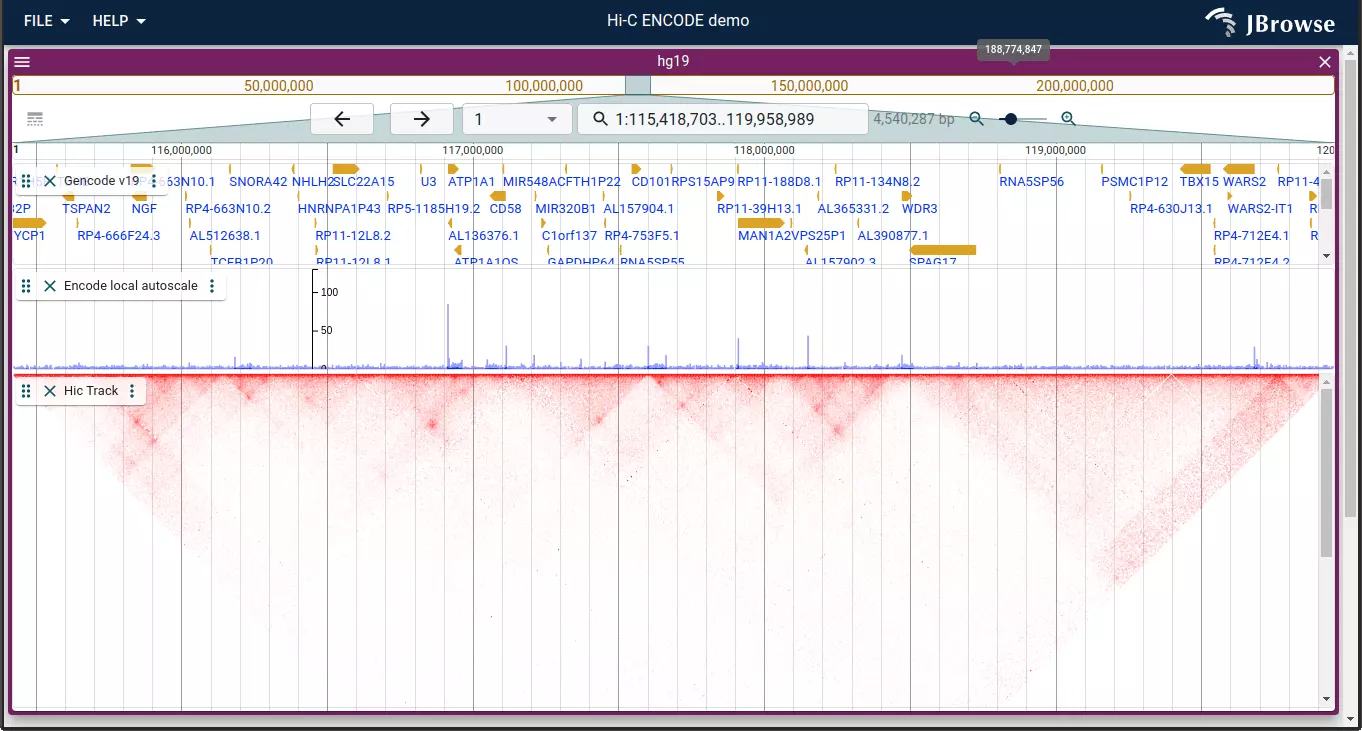
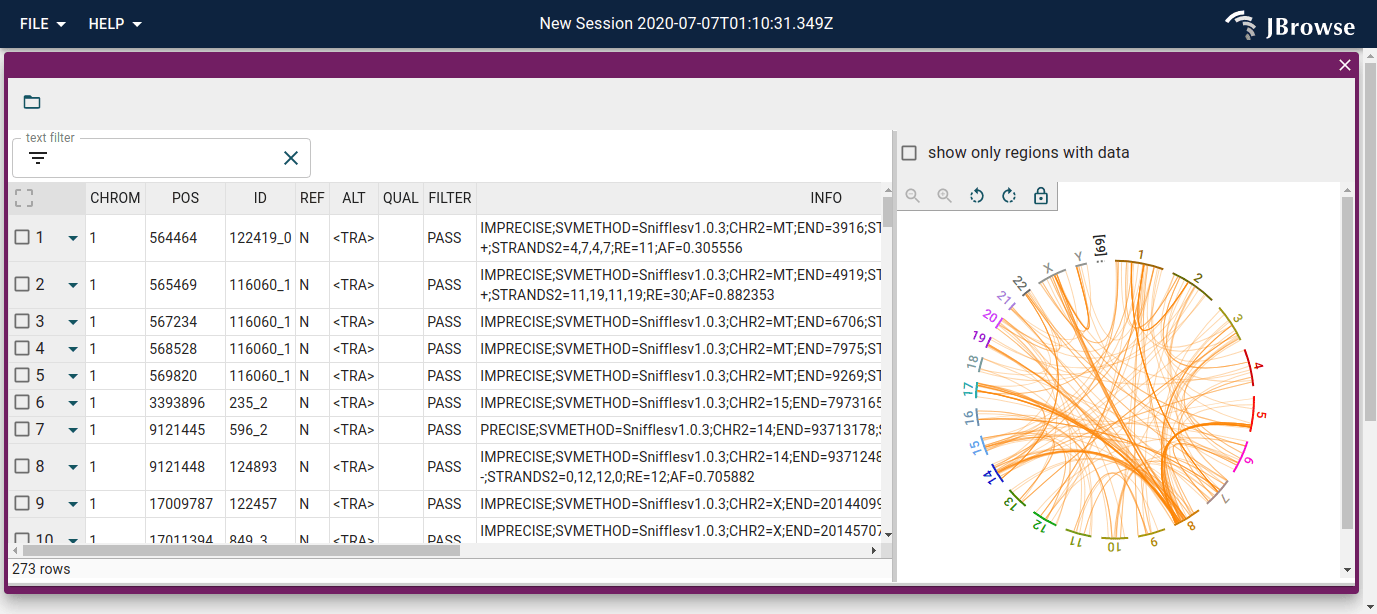
- Improved structural variant and comparative genomics visualization with linear, circular, dotplot, and synteny views
- Support for many common data types including BAM, CRAM, tabix indexed VCF, GFF, BED, BigBed, BigWig, and several specialized formats
- Endless extensibility with a plugin ecosytem which can add additional view types, track types, data adapters, and more!
- See a summary of new features and a comparison to JBrowse 1
Citation
We at the JBrowse Consortium are working to make JBrowse a pluggable, open-source computational platform for integrating many kinds of biological data from many different places.
Research citations are one of the main metrics the consortium uses to demonstrate our relevance and utility when applying for funding to continue our work. If you use JBrowse in research that you publish, please cite the most recent JBrowse paper:
JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biology (2023). https://doi.org/10.1186/s13059-023-02914-zFunding and Collaboration
JBrowse development is supported by the US National Institutes of Health (U41 HG003751), The Chan Zuckerberg Initiative, The Ontario Institute for Cancer Research (OICR), and the University of California, Berkeley.