JBrowse web setup using the GUI
In order to display your data, JBrowse 2 needs to know about the reference genome for your organism of interest and needs to have tracks created that reference your data sources. This guide will show you how to set those up using JBrowse 2's graphical configuration editing.
You can also do this configuration with JBrowse CLI. See here.
Pre-requisites
This tutorial requires having the following software installed
Starting JBrowse 2 admin server
The JBrowse CLI contains a tool called admin-server. This will act as a web
server for JBrowse 2 and will write any changes made in JBrowse 2 to a config
file.
Note The admin-server is meant to be used temporarily for configuration, not
in production.
The admin-server launches an instance of JBrowse 2 in "admin mode", which then
lets you:
- Add and edit assemblies with the "Assembly manager"
- Add tracks and edit tracks
- Add and edit connections
All of these changes will be written by the server to the JBrowse config file
(usually config.json) located in the JBrowse instance. This is something that
can only be done while the admin-server is running, which again, is only
meant to be temporary!
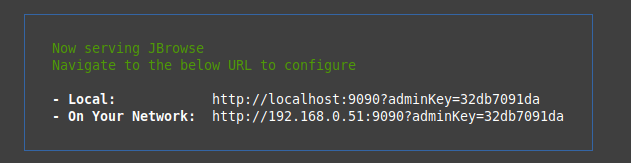
To start the admin-server, navigate into your JBrowse 2 directory and run:
## Start the admin-server
jbrowse admin-server
This will then generate a link that you can visit in your web browser:
Adding a genome assembly
A key first step in configuring a genome browser is adding an assembly to view.
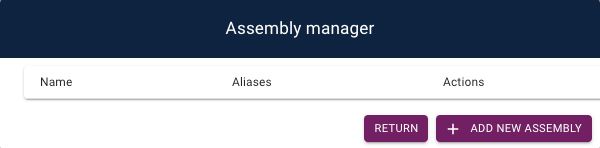
In order to do this, use the navigation bar to open up the Assembly Manager
(Admin > Open Assembly Manager).
This opens up a table which can be used to create, edit, and delete assemblies in your application:
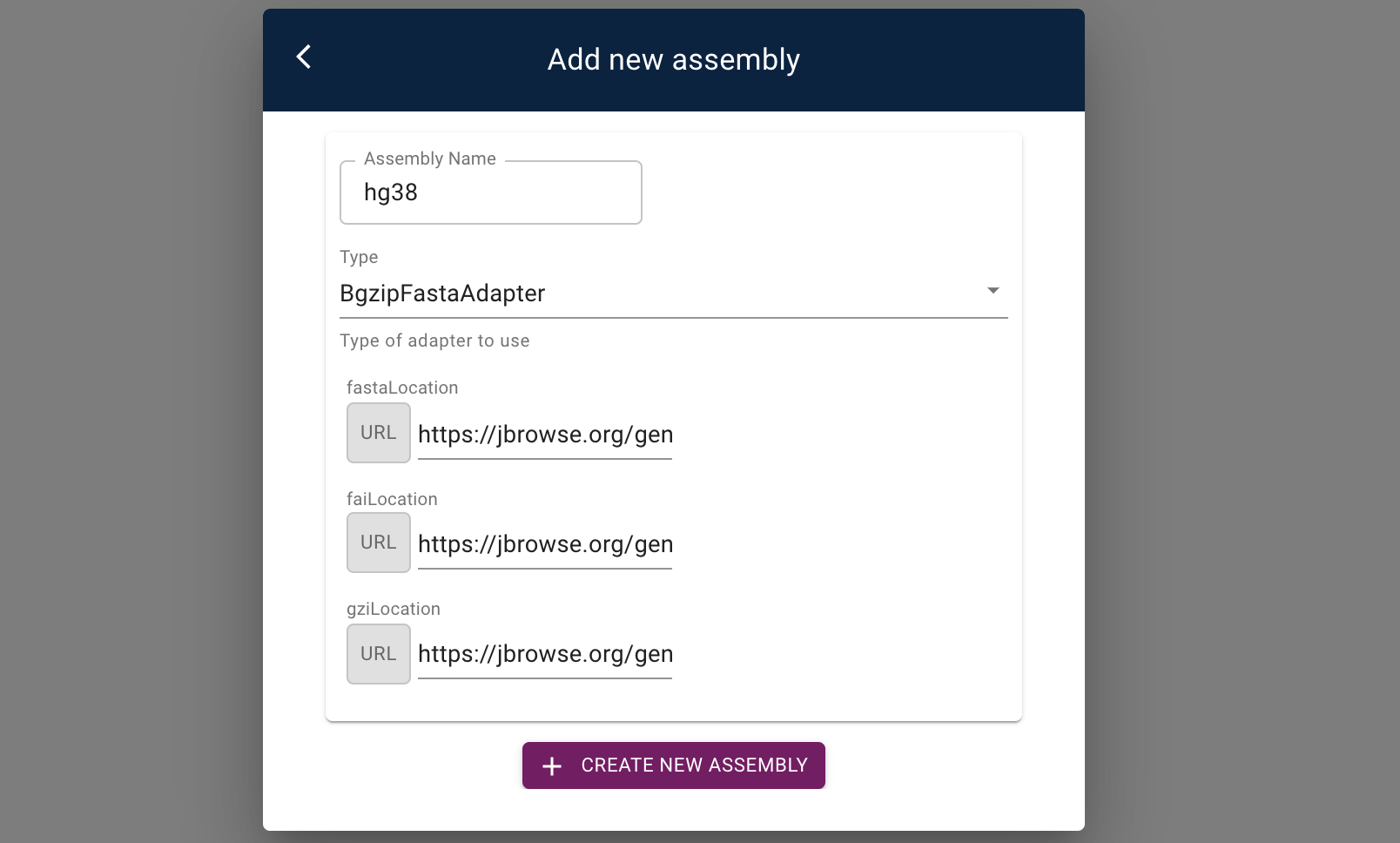
As an example, let's add the hg38 human reference genome to our JBrowse 2 application.
Press the "Add New Assembly" button, and enter the necessary information in the form:
- name:
hg38 - type:
BgzipFastaAdapter - fasta:
https://jbrowse.org/genomes/GRCh38/fasta/hg38.prefix.fa.gz - fasta index:
https://jbrowse.org/genomes/GRCh38/fasta/hg38.prefix.fa.gz.fai - gzi:
https://jbrowse.org/genomes/GRCh38/fasta/hg38.prefix.fa.gz.gzi
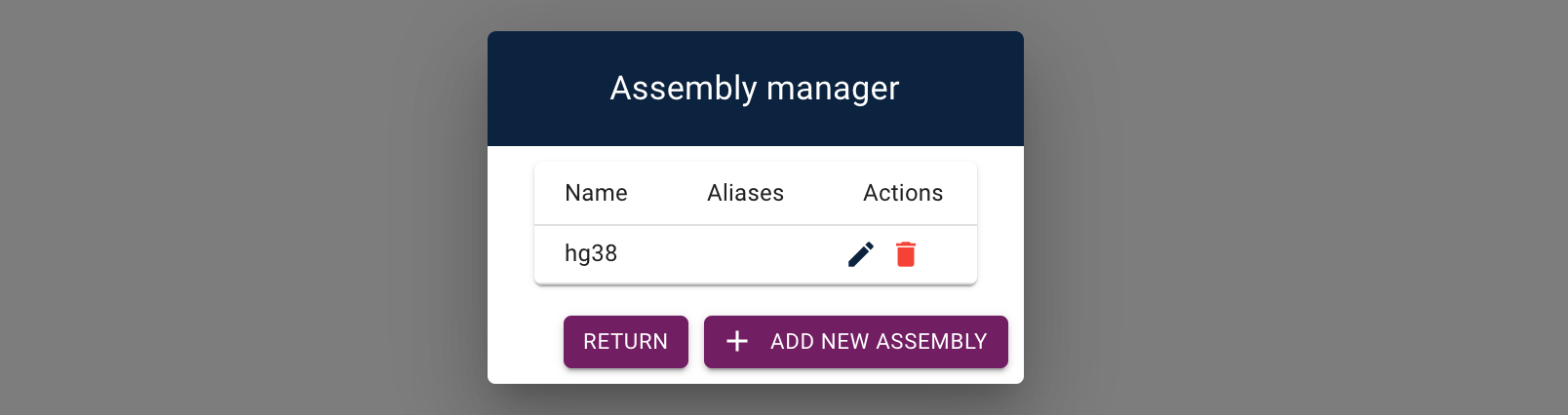
Click on "Create New Assembly". Great, we've added an assembly! We can see that we have successfully added the hg38 assembly:
The assembly can be edited or deleted, but for now we will return to the application.
Editing a genome assembly
After you've added a genome assembly, you can use the pencil icon button in the Assembly manager to edit that assembly. You can also delete assemblies from the assembly manager.
Adding a track
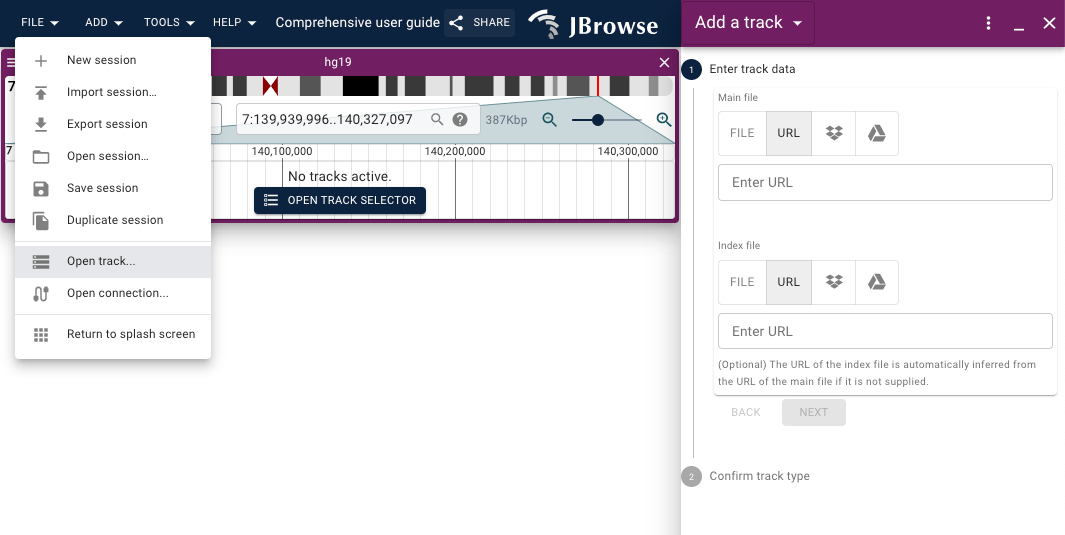
To add a new track or connection, you can use the menu bar in the app to open
the form for adding a track File > Open Track:
Alternatively, you can use the action button (circular "+") inside the track selector to access the "Add track" form:
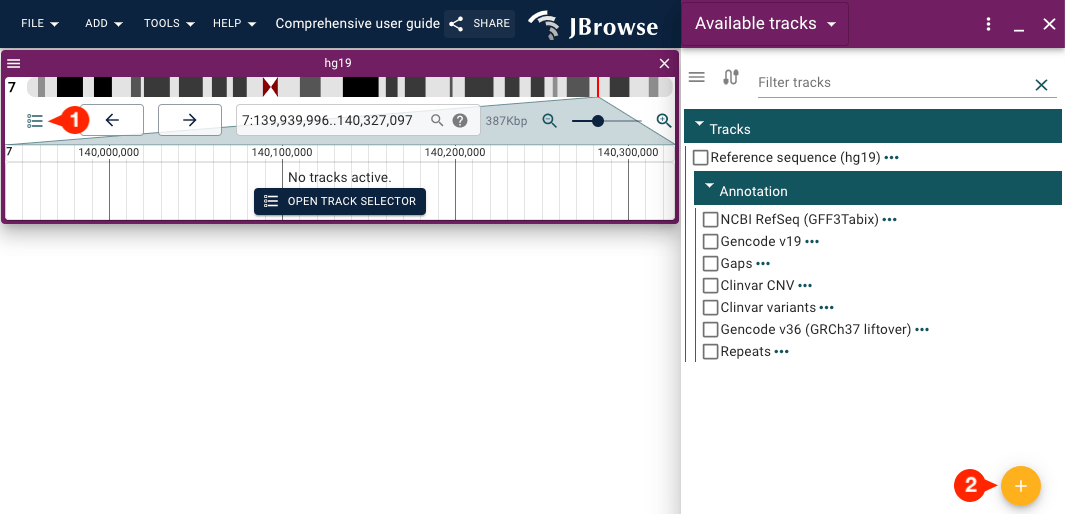
In the "Add track" form, you can provide a URL or select a local file to load.
The following file formats are supported in core JBrowse 2:
- CRAM
- BAM
- htsget
- VCF (Tabix-indexed)
- GFF3 (Tabix-indexed)
- BED (Tabix-indexed)
- BigBed
- BigWig
- JBrowse 1 nested containment lists (NCLists)
- plain text VCF, BED, CSV, TSV, BEDPE, STAR-fusion output (tabular formats)
- PAF (synteny/dotplot)
- Indexed FASTA/BGZip indexed FASTA
- 2bit
- .hic (Hi-C contact matrix visualization)
Additional data formats can be supported via plugins; checkout the plugin store.
For tabix files, TBI or CSI indexes are allowed. CSI or BAI is allowed for BAM. Only CRAI is allowed for CRAM. The index will be inferred for BAI or TBI files as e.g. filename+'.bai'. If it is different from this, make sure to specify the index file explicitly.
Editing a track
First, open a Linear Genome View using the navigation bar
(File > Add > Linear Genome View), and click on the "Select Tracks" button.
The configuration settings are accessible by clicking on the ellipses by each track.
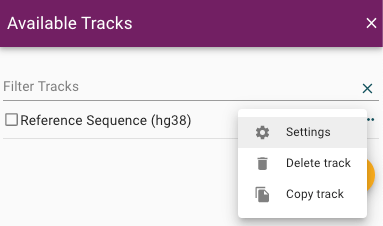
Open the configuration editor for the track by clicking on the "Settings" button shown above. You can use the configuration editor to live-edit any configurable value for a given track.
Setting a default session
It is also possible to use the graphical admin server to set the default session
of your JBrowse 2 instance. This is the session that will appear when JBrowse 2
is first visited. To do so, open the form to set the default session
(Admin > Set default session):
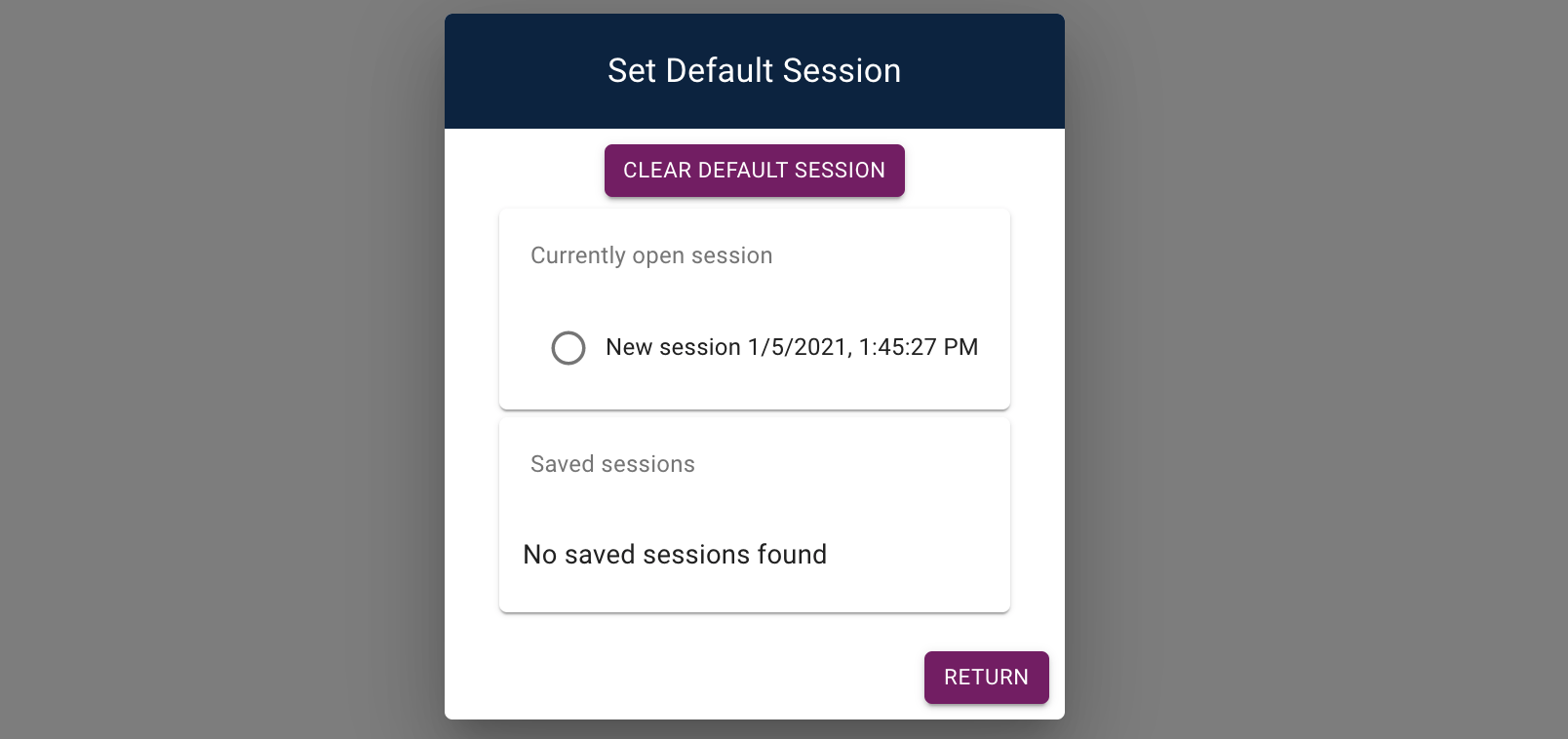
You can use the form to clear your default session, select the currently open session, or any of your previously saved sessions.
Additional resources
There are a number of additional features for configuring JBrowse 2. Make sure to refer to the config guide for more topics.
Conclusion
This guide showed how to launch the admin-server in the JBrowse CLI to perform
graphical configuration of your application. Specifically, we looked at how to
access and use the assembly manager, as well as how to access the configuration
editor for tracks. Importantly, all tracks have different configuration options
available in the configuration editor.
Make sure to take a look at any tracks you add to JBrowse 2 that you might want to further configure!