BCC2020 - Plugin development course
Welcome to the BCC2020 plugin development course
This will cover, starting from scratch, how we can develop a JBrowse 2 plugin
Here is a video recording of the presentation that we made
How to write a JBrowse 2 plugin. BCC2020 West Training on Vimeo
You can also follow along with the text presented below
Pre-requisites
Please have the following
- yarn, installed via https://classic.yarnpkg.com/en/docs/install/#debian-stable
or
npm install -g yarn - node v10 or greater, installed via https://github.com/nodesource/distributions#installation-instructions
These links above provide more reliable installation than the apt repositories version, but you can try to install these however you feel comfortable
Install CLI tools
To start, we will install the jbrowse-cli
sudo npm install -g @jbrowse/cli
Depending on your setup you might need to use sudo for this. We will use the CLI to download the latest release from github, this is a convenience tool that saves us some steps from manually downloading it
Create a jbrowse 2 production instance
cd /var/www/html
sudo jbrowse create myfolder
This will create a jbrowse2 instance in a folder named myfolder. We can now visit http://localhost/myfolder and see that this gives us a message that jbrowse2 is not yet configured
Outline for our plugins
Now that we have initialized an instance, we will look at how to use plugins
We are going to create several plugins for this tutorial
- Downloading data from the UCSC API
- Creating custom drawing code
- A template for a custom view type
UCSC API data adapter
Let's start with creating a plugin that accesses the UCSC REST API. See https://genome.ucsc.edu/goldenPath/help/api.html for docs
We will clone a working version of this plugin for brevity and analyze it
The development and build process for plugins has changed since this tutorial was created. See https://github.com/GMOD/jbrowse-plugin-template#readme for current instructions.
cd ~/
git clone https://github.com/cmdcolin/jbrowse-plugin-ucsc-api
cd jbrowse-plugin-ucsc-api
yarn
yarn develop --port 9001
This will build the plugin and serve it with a webpack-dev-server on port 9001
We can then load our in-development plugin with our production version of jbrowse http://localhost/myfolder/?config=http://localhost:9001/config_ucsc_api.json
If you had a github clone of jbrowse-components, with the products/jbrowse-web started, you could do this with http://localhost:3000/?config=http://localhost:9001/config_ucsc_api.json instead
Analysis of the UCSC REST API plugin
Notes about the plugin:
- we run our plugin development server on a custom port. This is a webpack-dev-server for the plugin code
- the config we are pointing at is here https://github.com/cmdcolin/jbrowse-plugin-ucsc-api/blob/master/assets/config_ucsc_api.json and we can see it is basically resolving to a plugin.js file at a CDN, which can be the final built output or the webpack-dev-server served version
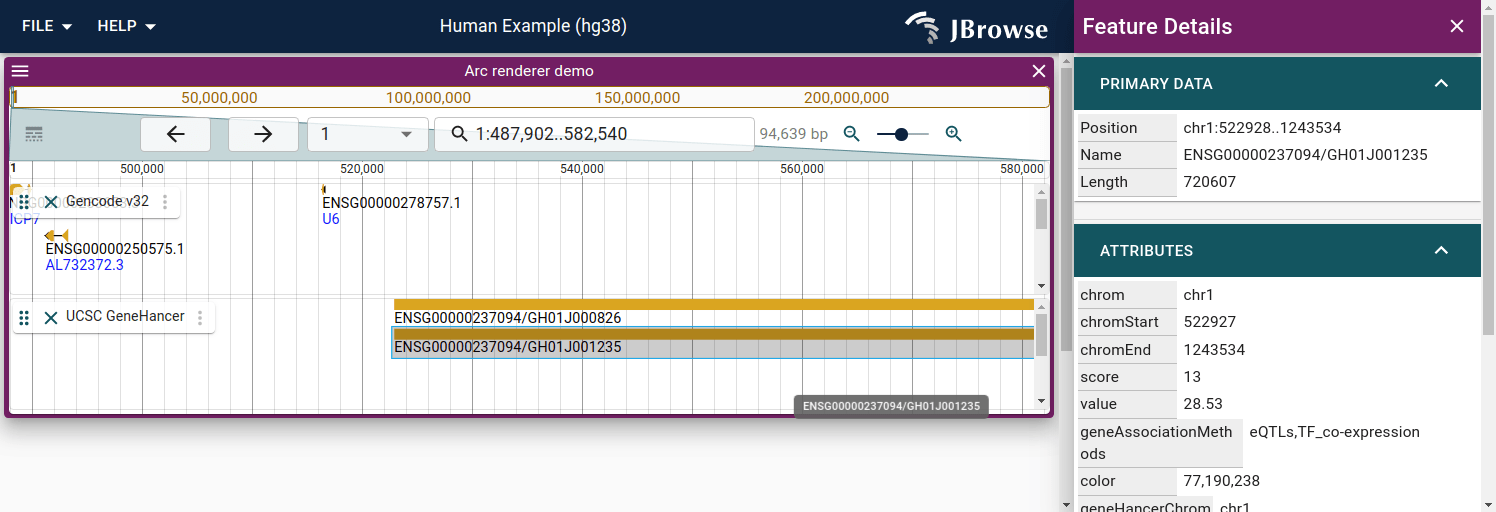
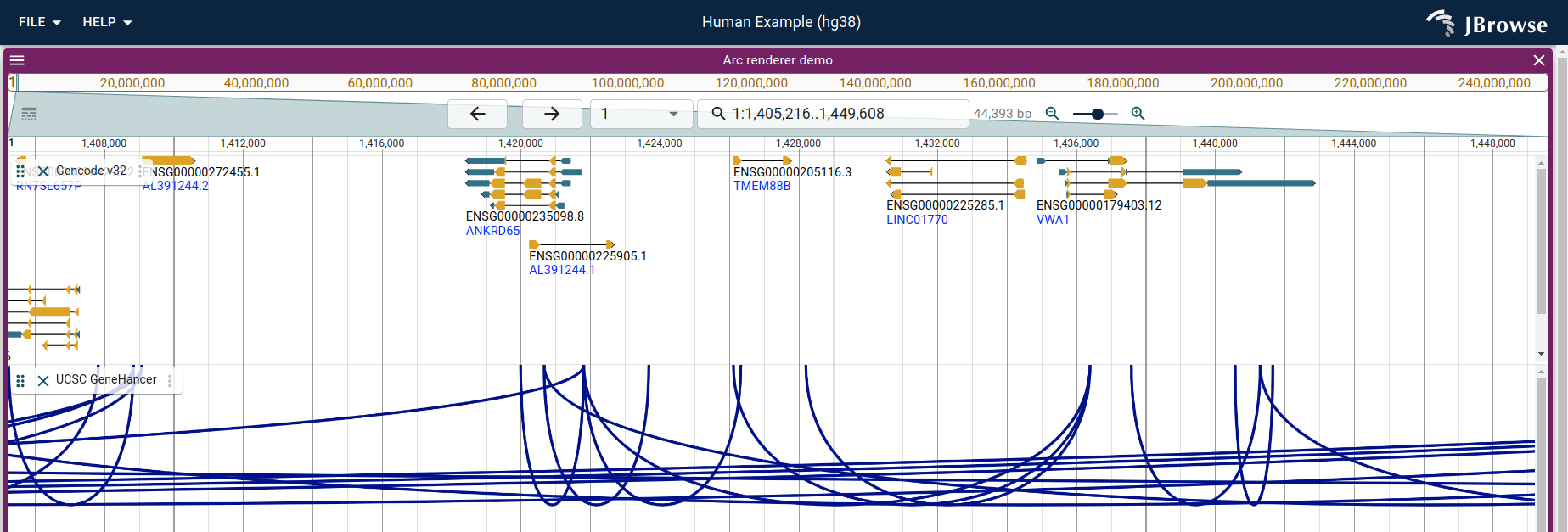
Combining the UCSC API plugin with a custom renderer
Interaction data is often displayed using arcs to connect enhancer to gene. We will create a custom renderer to illustrate this
But what is a renderer? It is code that performs drawing. See the renderer docs here for more details on creating renderers
Let's clone a working arc renderer plugin
git clone https://github.com/cmdcolin/jbrowse-plugin-arc-renderer
cd jbrowse-plugin-arc-renderer
yarn
yarn develop --port 9000
This will start the plugin for the arc renderer on port 9000. Now, we will keep the UCSC API plugin running on port 9001, and then visit
http://localhost/myfolder/?config=http://localhost:9000/config_arc_renderer.json
This will load the following file https://github.com/cmdcolin/jbrowse-plugin-arc-renderer/blob/master/assets/config_arc_renderer.json
This loads both the UCSCPlugin and the ArcRendererPlugin at the same time, and renders the UCSC GeneHancer interactions as arcs
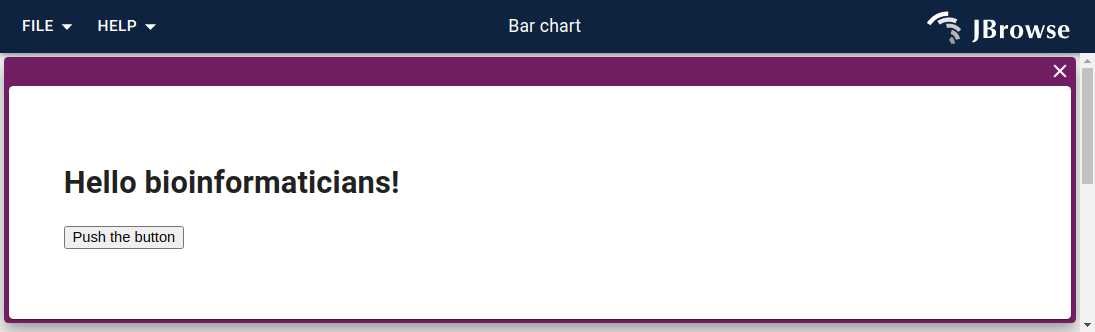
Making custom view types with plugins
Many new things are possible by making completely custom view types in JBrowse 2
Plugins can basically register a new view type that is a ReactComponent without much else, allowing integration of diverse other view types that are not really constrained at all
Here is a template we can work from
https://github.com/cmdcolin/jbrowse-plugin-barchart-view
Here is a silly example with a custom Hello world view type. I started this as a "bar chart" concept but only got as far as making it say hello world.
Looking at the code, it is fairly simple and demonstrates that we can basically have any sort of ReactComponent rendered into our view. That means we could have a gene expression heatmap, barchart, get charts dynamically from an R server side component, make a graph genome, etc. The ideas are endless! And we can make it interact with other views!
Debugging your plugins
The examples allowed us to get us quickly setup
In your daily work we encourage you to clone the jbrowse-components repo and run off a dev version of jbrowse 2 e.g. follow the steps below
git clone https://github.com/gmod/jbrowse-components
cd jbrowse-components
yarn
cd products/jbrowse-web
yarn start
Then point your dev version of jbrowse 2 at the dev version of your plugin http://localhost:3000/?config=http://localhost:9001/config_ucsc_api.json
A major benefit to using a github clone of jbrowse 2 compared with the version
from jbrowse create myfolder is that you will get better stack traces
Loading a genome and using plugins in production
In the previous examples, we used our sample data from a plugins preconfigured configs
Let's start to create a production instance of JBrowse. We can use the jbrowse CLI tools to load some data. We will refer to an existing genome fasta file on the web, and a refname alias map (tab separated association of chr1->1, etc.)
cd /var/www/html/myfolder
jbrowse add-assembly https://jbrowse.org/genomes/hg19/fasta/hg19.fa.gz --refNameAliases https://jbrowse.org/genomes/hg19/hg19_aliases.txt
Then we could add a gene track
jbrowse add-track https://s3.amazonaws.com/jbrowse.org/genomes/hg19/GRCh37_latest_genomic.sort.gff.gz --config '{"renderer": {"type": "SvgFeatureRenderer"}}'
Or a track from UCSC
jbrowse add-track https://jbrowse.org/genomes/hg19/knownGene.bb --config '{"renderer": {"type": "SvgFeatureRenderer"} }'
Now visit http://localhost/myfolder and open up a new linear genome view with the hg19 assembly and you should see these tracks.
Now you can edit your config to contain runtime plugins and use this in production
Conclusion
This is an initial look into jbrowse 2 plugin development. I strongly encourage reading the developer guide in the main documentation for more info, and let us know if you have any feedback or questions. Thanks!