SV inspector view
The Structural Variant (SV) inspector is a "workflow" that is designed to help users inspect structural variant calls.
We can start the SV inspector by launching it from the App level menu bar
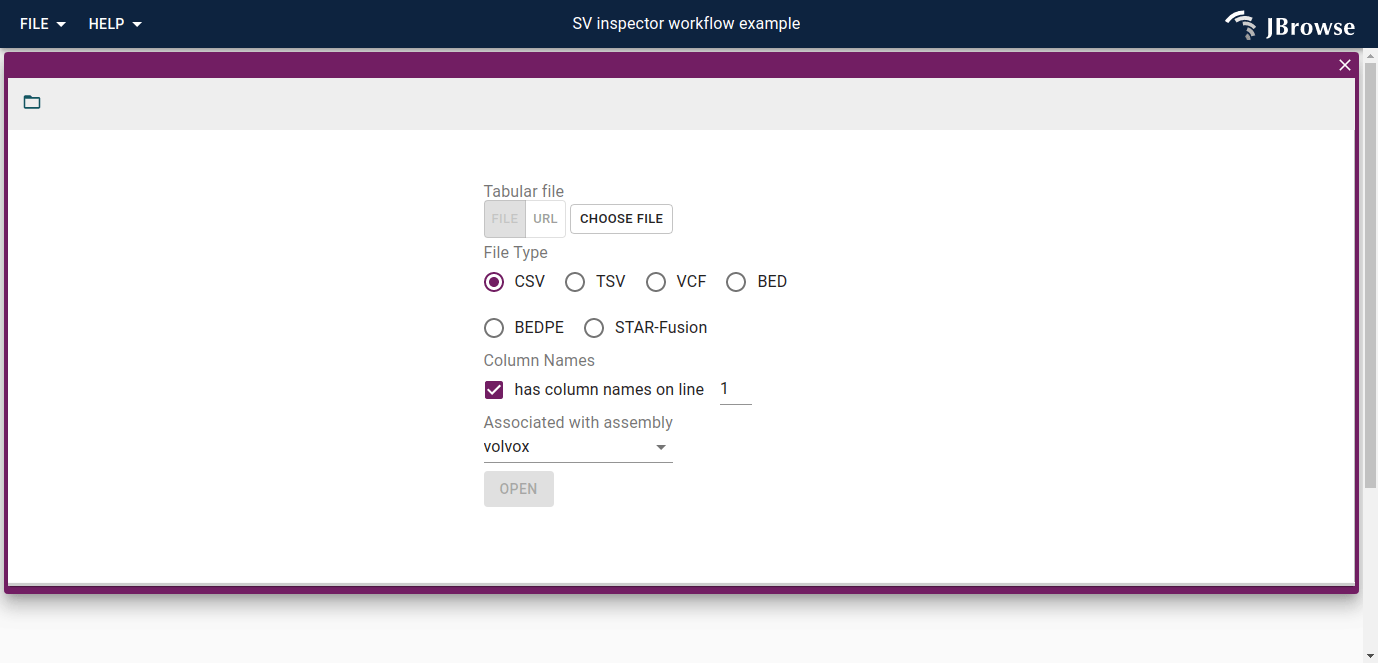
This will bring up an "import form" that asks you for your SV evidence.
The following formats are supported:
- CSV, TSV
- VCF or VCF.gz (plain text VCF, or (b)gzipped VCF)
- BED, BEDPE
- STAR-fusion result file
Sources of data for SV inspector
The SV inspector currently is designed for viewing <TRA> and breakend type
entries.
Examples of variant callers that produce a VCF that can be used with the SV inspector:
Short read based:
- Manta
- Delly
- Lumpy
Long read based
- pbsv
- Sniffles
Example SV inspector workflow
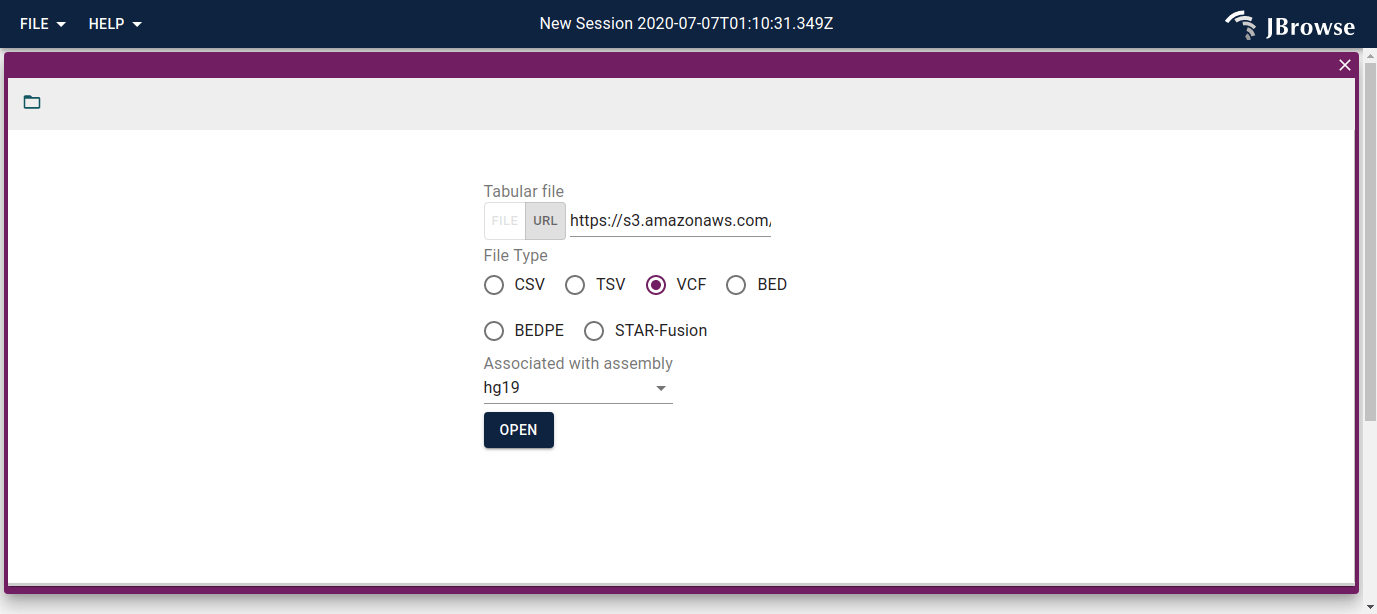
We can start the SV inspector workflow by opening up this file containing translocation events called from a breast cancer cell line SKBR3, based on these published data.
Example VCF for use in the SV inspector
Copy this URL and paste it into the import form and select hg19:
SV inspector results
After loading the user's requested file, you will have a tabular view with each row representing a row of the file you opened, along with a whole-genome overview of the SVs on the right
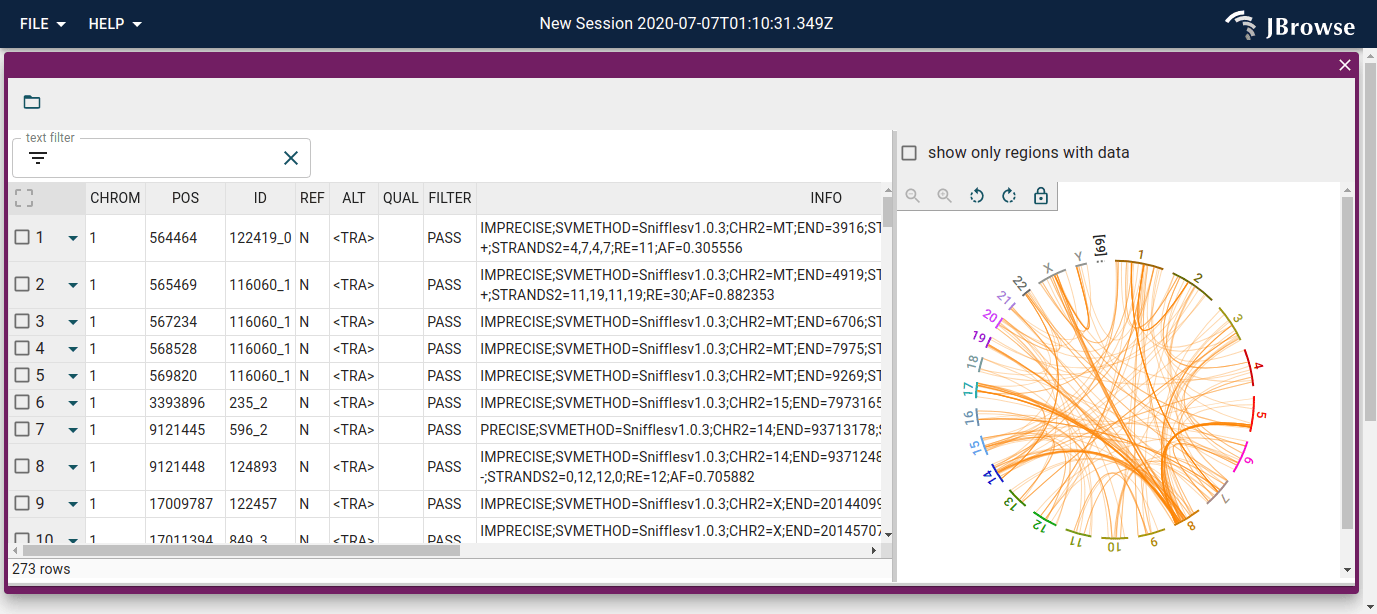
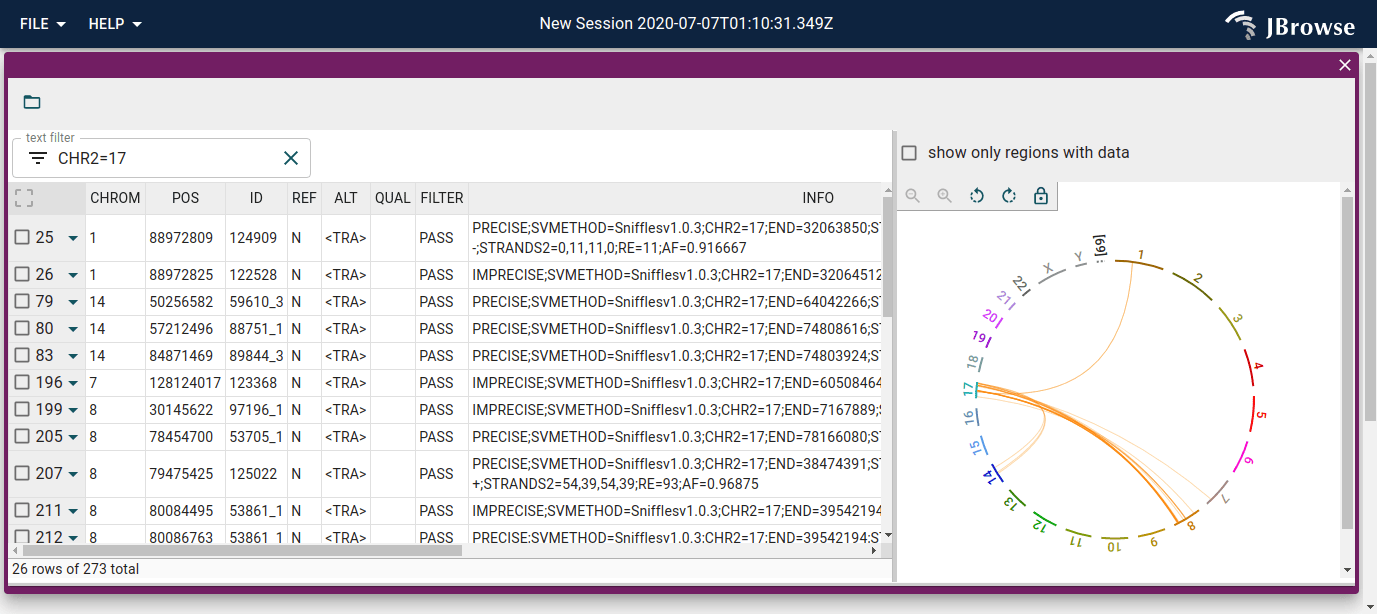
We can search and filter the table, with filtering and searching being reflected in the circular view as well.
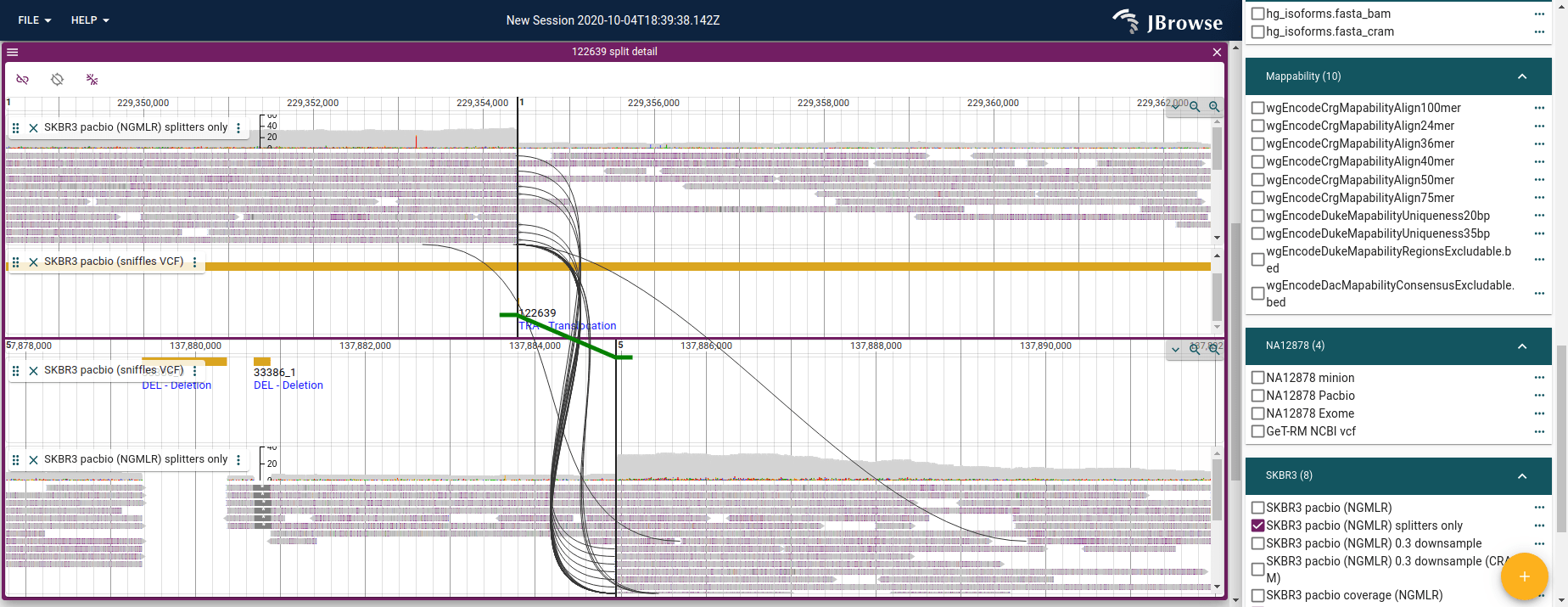
Launching breakpoint split view
By clicking on the features in the circular view, or clicking on the triangle drop-down on the leftmost column of the table, we can dynamically launch a new view of the data that is called the "split view" or the "breakpoint split view"
This allows us to inspect the breakpoints of the structural variant, and compare each side to the alignments.