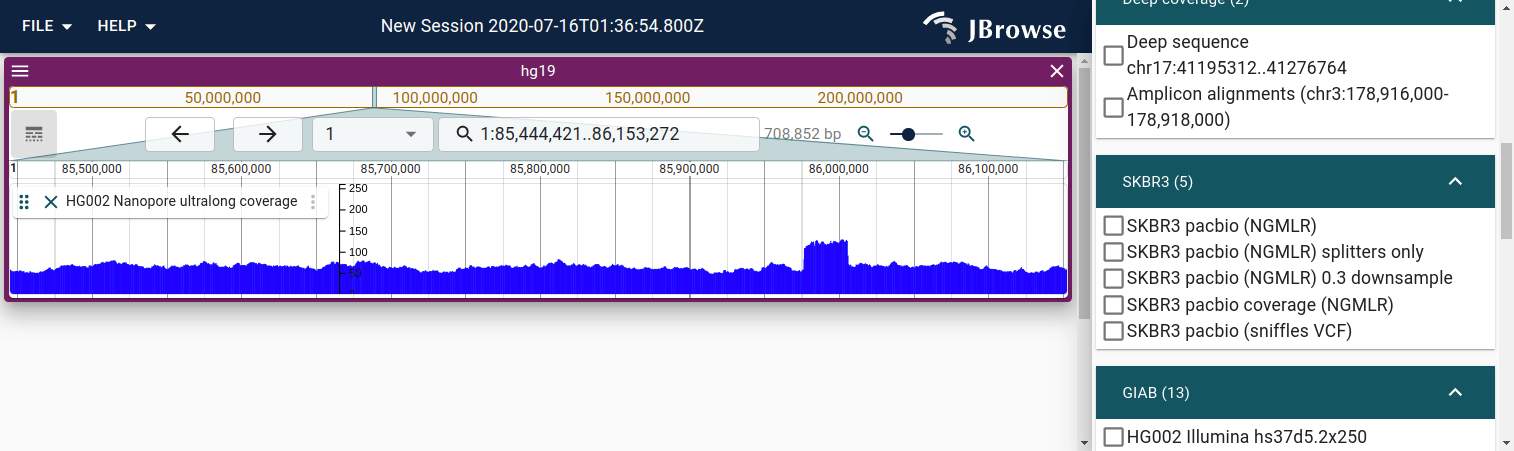
Quantitative tracks
Visualizing genome stopTokens, whether it is read depth-of-coverage or other stopToken, can often be done by using BigWig or other quantitative feature files.
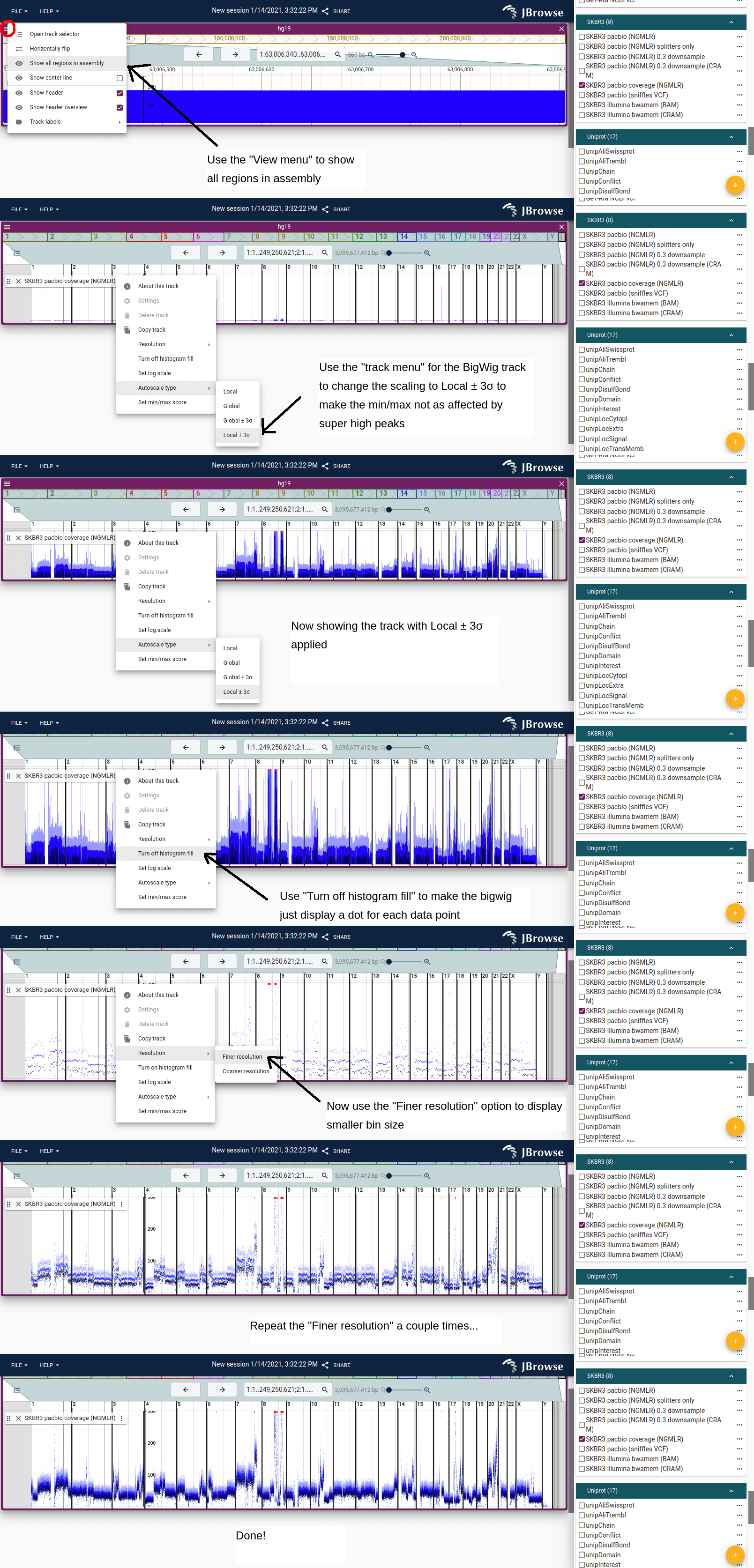
Viewing whole-genome coverage for profiling CNV
You can refine the resolution of BigWig tracks, and view whole genome coverage to get detailed global views of CNV, for example from whole-genome coverage profiling.
Here is a short picture guide to setup a whole-genome view of a BigWig for CNV coverage visualization:
- Open your BigWig track
- Go to the view menu and select "Show all assembly regions"
- Adjust the "Autoscale type" to your liking, the new options for "Local +/- 3sd" allows the autoscaling to avoid outliers
- Go to the track menu and select "Turn off histogram fill", which then shows only a dot for each point on the graph
- Go to the track menu and select "Resolution->Finer resolution" a couple times until resolution looks nice
Note
All tracks have a drag handle on the bottom, which you can drag down to make the track taller.