JBrowse web setup using the CLI
Prerequisites
- Node.js 18+. Note: we recommend not using
aptto install Node.js, it often installs old versions. Good alternatives include NodeSource or NVM. - Samtools installed e.g.
sudo apt install samtoolsorbrew install samtools, used for creating FASTA index and BAM/CRAM processing for creating tabix GFF - tabix installed e.g.
sudo apt install tabixorbrew install htslib, used for creating tabix indexes for BED/VCF/GFF files
Installing the JBrowse CLI
The JBrowse CLI can help perform many tasks to help you manage JBrowse 2, such as:
- create a new instance of JBrowse 2 automatically
- update an existing instance of JBrowse 2 with the latest released version
- configure your JBrowse 2 instance
To globally install the JBrowse CLI, run
npm install -g @jbrowse/cli
After running this command you can then test the installation with
jbrowse --version
which will output the current version of the JBrowse CLI.
If you can't or don't want to globally install the JBrowse CLI, you can also use
the npx command, which
is included with Node.js, to run JBrowse CLI without installing it. Simply
replace jbrowse with npx @jbrowse/cli in any command, e.g.
npx @jbrowse/cli --version
Another way which involves downloading a single file bundle is to use
wget https://unpkg.com/@jbrowse/cli/bundle/index.js -O jbrowse
chmod +x jbrowse
./jbrowse --help
Using jbrowse create to download JBrowse 2
In the directory where you would like to download JBrowse 2, run
jbrowse create jbrowse2
This fetches the latest version of jbrowse-web and unzips it to a folder named "jbrowse2". You can name the folder anything you want to, it is just a folder containing html, css, and js files. It is not a fancy installation
Also note: as an alternative to the jbrowse create command, you can manually download the jbrowse-web zip from https://github.com/GMOD/jbrowse-components/releases
Checking the download
The directory where you downloaded JBrowse should look something like this:
jbrowse2/
- asset-manifest.json
- favicon.ico
- index.html
- manifest.json
- robots.txt
- static/
- test_data/
- version.txt
Running JBrowse 2
JBrowse 2 requires a web server to run. It won't work if you try to directly
open the index.html in your web browser.
Oftentimes, you may put the folder on a web server in the static html folder e.g. /var/www/html/jbrowse2/ once in place, you can then visit http://yourserver/jbrowse2
You could also use a simple server to check that JBrowse 2 has been downloaded properly. Run
cd jbrowse2/
npx serve .
# or
npx serve -S . # if you want to refer to symlinked data later on
which will start a web server in our JBrowse 2 directory.
Navigate to the location specified in the CLI's output (likely
http://localhost:3000).
Your page should look something like this:
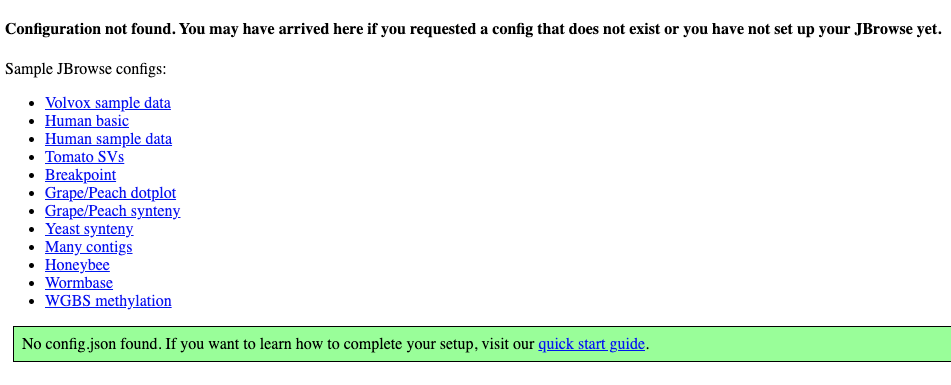
Click on the sample config to see JBrowse 2 running with a demo configuration. It should look like this:
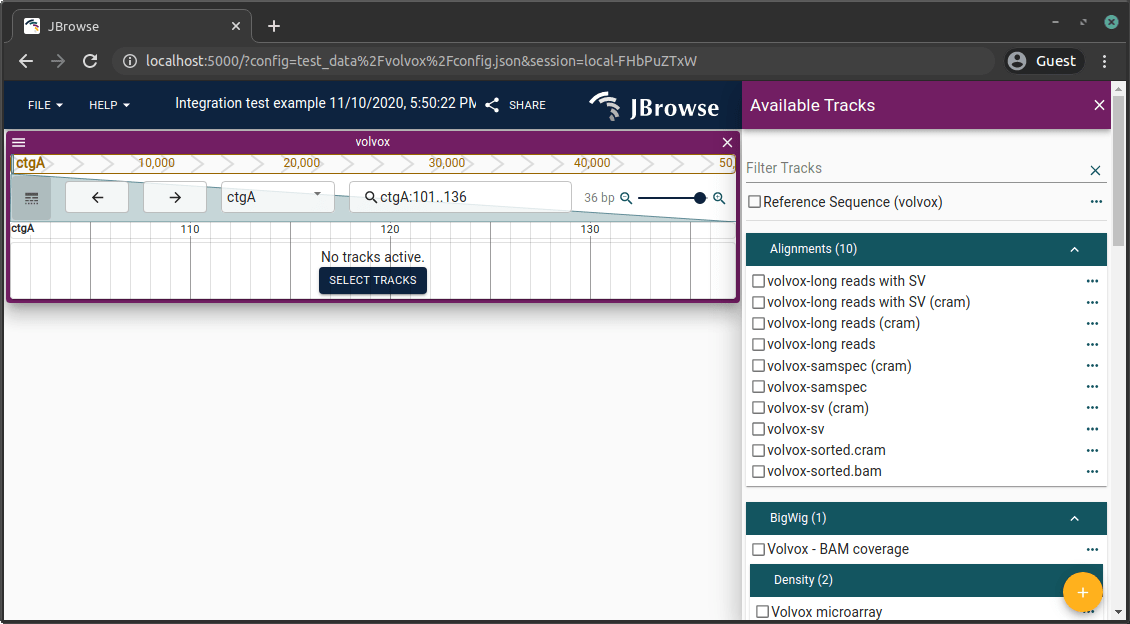
Congratulations! You're running JBrowse 2.
Adding tracks
Now that JBrowse 2 is set up, you can configure it with your own genomes and tracks.
Adding a genome assembly in FASTA format
The first step to creating a jbrowse config is to load a genome assembly. This
is normally in FASTA format, and we will start by creating a "FASTA index" with
samtools:
samtools faidx genome.fa
jbrowse add-assembly genome.fa --load copy --out /var/www/html/jbrowse/
This will output a configuration snippet to a file named /var/www/html/jbrowse/config.json if it does not already exist, or append a new assembly to that config file if it does exist. It will also copy genome.fa and genome.fa.fai to the /var/www/html/jbrowse/ folder because we used --load copy. If you wanted to symlink instead, can use --load symlink
JBrowse 2 also supports other assembly file formats, including bgzip-compressed indexed FASTA, and 2bit files.
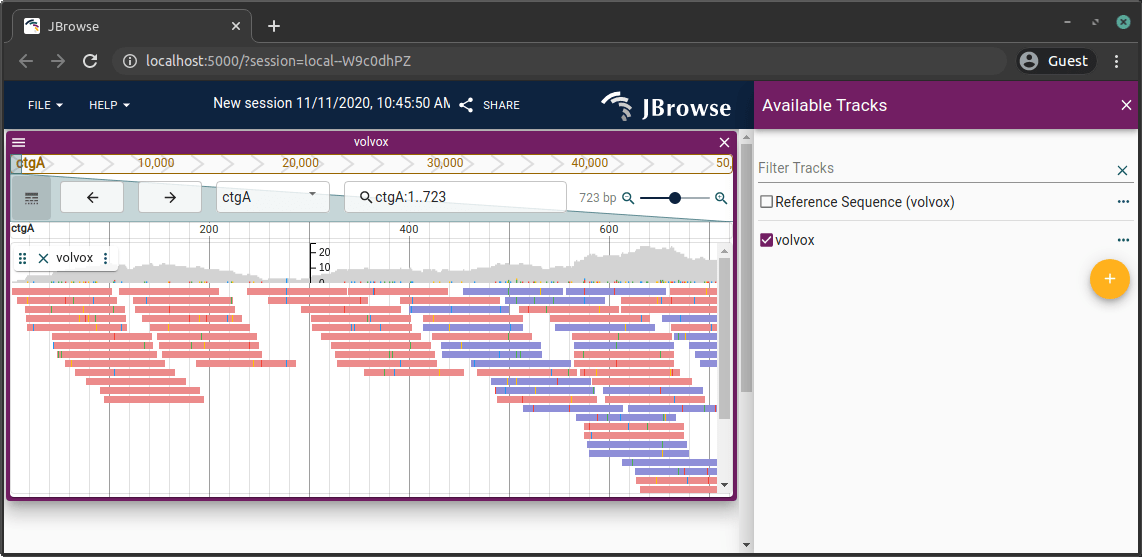
Adding a BAM or CRAM track
For this example, we will use a BAM file to add an alignments track. As with assemblies, you can add a track using local files or remote locations of your files.
samtools index file.bam
jbrowse add-track file.bam --load copy --out /var/www/html/jbrowse
samtools index file.cram
jbrowse add-track file.cram --load copy --out /var/www/html/jbrowse
This will add a track configuration entry to /var/www/html/jbrowse/config.json
and copy the files into the folder as well. If you use --load symlink, it can
symlink the files instead. To see more options for adding the track, such as
specifying a name, run jbrowse add-track --help.
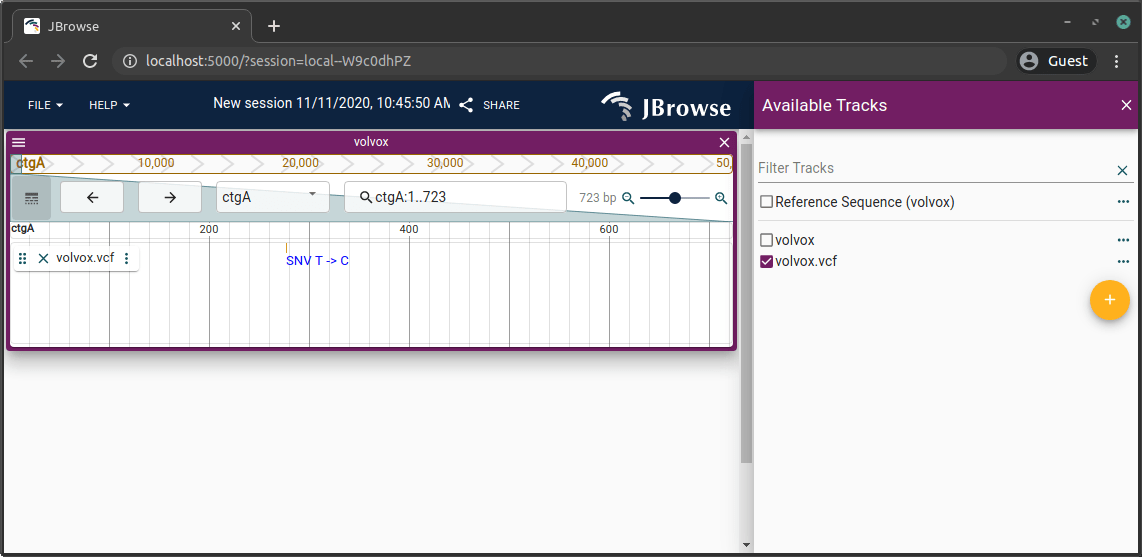
If you have JBrowse 2 running as described in the JBrowse web quickstart, you can refresh the page and an add a linear genome view of the volvox assembly. Then open track selector, and you will see the alignments track.
Adding a VCF track
Adding a variant track is similar to adding an alignments track. For this
example, we will use a VCF file for the track. JBrowse 2 expects VCFs to be
compressed with bgzip and tabix indexed.
To add the track, run
bgzip file.vcf
tabix file.vcf.gz
jbrowse add-track file.vcf.gz --load copy --out /var/www/html/jbrowse
Note if you get errors about your VCF file not being sorted when using tabix, you can use bcftools to sort your VCF.
bcftools sort file.vcf > file.sorted.vcf
bgzip file.sorted.vcf
tabix file.sorted.vcf.gz
You can also bgzip and index with the bcftools tool.
bcftools view volvox.vcf --output-type z > volvox.vcf.gz
rm volvox.vcf
bcftools index --tbi volvox.vcf.gz
For more info about bgzip, tabix, and bcftools, see
https://www.htslib.org/.
Adding a BigWig/BigBed track
Probably one of the most simple track types to load is a BigWig/BigBed file since it does not have any external index file, it is just a single file.
jbrowse add-track file.bw --load copy --out /var/www/html/jbrowse
Adding a GFF3 file with GFF3Tabix
jbrowse sort-gff yourfile.gff | bgzip > yourfile.sorted.gff.gz
tabix yourfile.sorted.gff.gz
jbrowse add-track yourfile.sorted.gff.gz --load copy
Note: the jbrowse sort-gff command just automates the following shell command
(grep "^#" in.gff; grep -v "^#" in.gff | sort -t"`printf '\t'`" -k1,1 -k4,4n) > sorted.gff;
This command comes from the tabix documentation
Adding a synteny track from a PAF file
Loading synteny data makes use of all the previous functions we've used so far in this guide.
Here, we make use of the grape and peach genome assemblies, but replace with your own data if applicable.
Use minimap2 to create a PAF file from FASTA files:
## Use minimap2 to create a PAF from your assemblies
minimap2 grape.fa peach.fa > peach_vs_grape.paf
## add each assembly to jbrowse config
## the -n flag names the assemblies explicitly
jbrowse add-assembly grape.fa --load copy -n grape --out /var/www/html/jbrowse
jbrowse add-assembly peach.fa --load copy -n peach --out /var/www/html/jbrowse
Next, we'll load the synteny "track" from the PAF file.
Order matters here for the --assemblyNames parameter:
If minimap2 is run as minimap2 grape.fa peach.fa, then you need to load as
--assemblyNames peach,grape.
The order is reversed between the minimap2 and jbrowse tools.
jbrowse add-track peach_vs_grape.paf --assemblyNames peach,grape --load copy --out /var/www/html/jbrowse
Indexing feature names for searching
The final step of loading your JBrowse instance may include adding a "search index" so that you can search by genes or other features by their name or ID.
To do this we can use the jbrowse text-index command:
jbrowse text-index --out /var/www/html/jbrowse
This will index relevant track types e.g. any track with Gff3TabixAdapter (gene names and IDs) or VcfTabixAdapter (e.g. variant IDs). The command will print out a progress bar for each track that it is indexing.
This will also update your config.json so that after it completes, you can
type a gene name into the "search box" in the linear genome view or other views
and quickly navigate to genes by gene name.
See the text-index command docs for more info. Also see the FAQ entries for text searching
Conclusion
Now that you have JBrowse configured with an assembly and a couple of tracks, you can start customizing it further. Check out the rest of the docs for more information, especially the JBrowse CLI docs for more details on some of the steps shown here.
Miscellaneous tips
You can use --subDir to organize your data directory:
mkdir my_bams
## Copies .bam and .bai files to my_bams folder
jbrowse add-track myfile.bam --subDir my_bams --load copy --out /var/www/html/jbrowse
Upgrade JBrowse Web to the latest version
You can upgrade your JBrowse release to the latest version with:
# run this command in an existing jbrowse 2 installation
jbrowse upgrade
The above command downloads the latest jbrowse-web from github and unzips it into the current directory
Upgrade @jbrowse/cli to the latest
To upgrade the CLI tools, you can re-run the install command
npm install -g @jbrowse/cli
Output to a custom config file name
You can use filenames other than config.json, and put configs in subfolders of your jbrowse 2 installation too
jbrowse add-assembly mygenome.fa --out /path/to/my/jbrowse2/alt_config.json --load copy
This would then be accessible at e.g. http://localhost/jbrowse2/?config=subfolder/alt_config.json