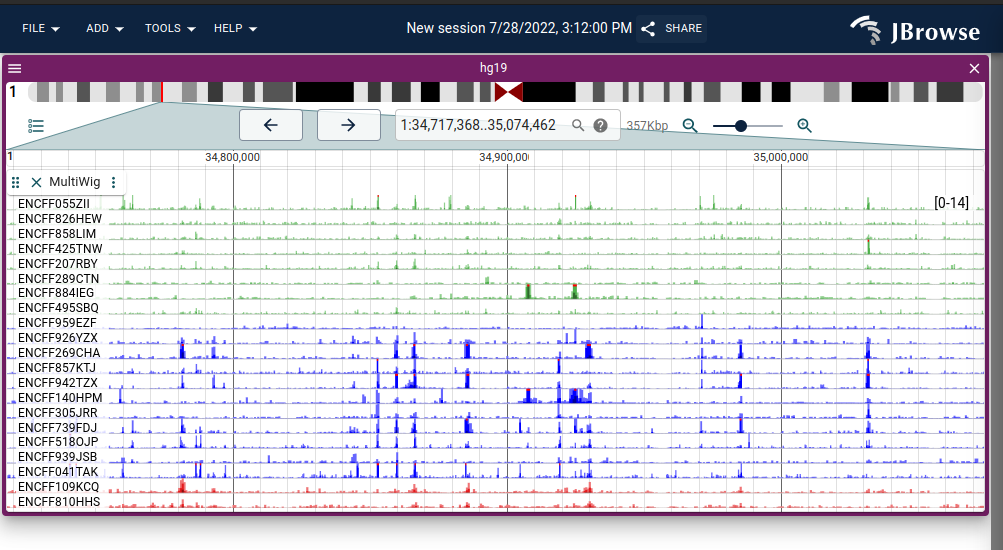
v2.2.2 Release
Hello all, this release contains a bugfix for the circular view and breakpoint split view with "TRA" type features not being displayed. See release notes below for a couple other small bugfixes and enhancements.
Downloads
To install JBrowse 2 for the web, you can download the link above, or you can use the JBrowse CLI to automatically download the latest version. See the JBrowse web quick start for more details.
2.2.2 (2022-12-06)
🚀 Enhancement
- Other
- #3350 Retain feature labels in compact display mode for SVG features, and allow keeping feature description without feature label (@cmdcolin)
- #3357 Allow holding shift key to create rubberband selection on LGV (@cmdcolin)
- #3363 Create remove-track CLI command (@cmdcolin)
- #3341 Add BEDPE adapter type (@cmdcolin)
core
🐛 Bug Fix
- Other
- #3377 Fix rendering and clicking synteny features when using MainThreadRpc (@cmdcolin)
- #3375 Fix
jbrowse upgradeCLI command overwriting config.json with --branch or --nightly options (@cmdcolin) - #3370 Fix error with SNPCoverage not rendering MM tag modifications in some cases (@cmdcolin)
- #3366 Fix ability to use LGV synteny track on inverted alignments (@cmdcolin)
- #3348 Fix for
breakpoint split view and circular view issues with
<TRA>type entries in v2.2.1 (@cmdcolin)
core
📝 Documentation
🏠 Internal
coretext-indexing


 Figure showing improved speed on short read alignments of the refactor (this
release) vs main (which was v2.1.7)
Figure showing improved speed on short read alignments of the refactor (this
release) vs main (which was v2.1.7)