This release adds a new option "Color by CDS" to color the reference sequence
track and CDS features in gene tracks by their reading frame
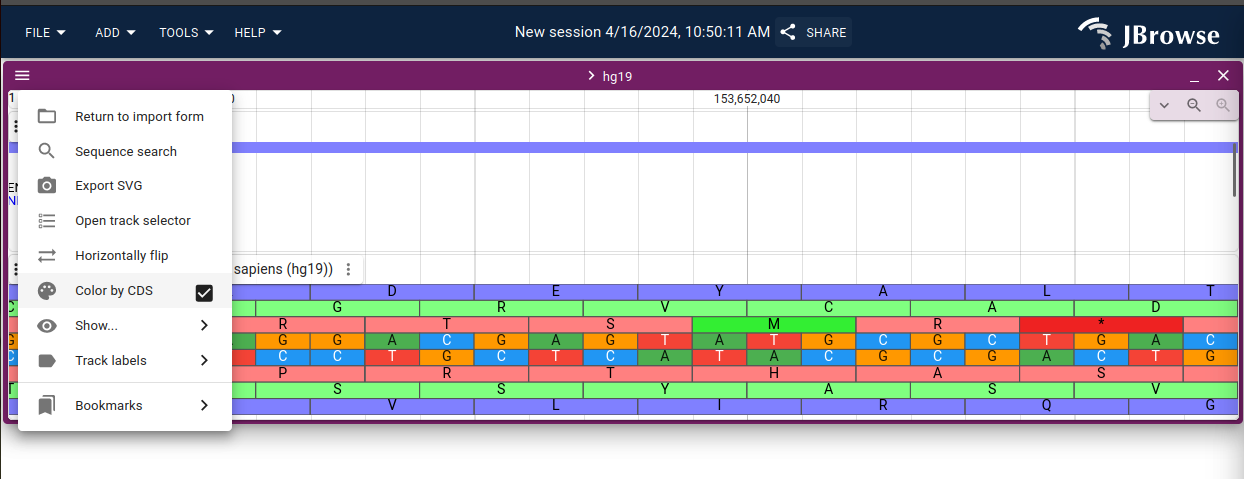
Screenshot showing the "Color by CDS" setting on the linear genome view menu
This release also adds new Hi-C coloring options, including a log-scale mode
that enhances visible patterns

There are also many other small fixes including the ability to highlight
multiple regions from the URL bar &highlight= option, faster zooming with the
zoom in/out buttons, and more
See release notes for details
Downloads
To install JBrowse 2 for the web, you can download the link above, or you can
use the JBrowse CLI to automatically download the latest version. See the
JBrowse web quick start for more
details.
yarn run v1.22.19 $ lerna-changelog --silent --silent --next-version 2.11.0
2.11.0 (2024-04-16)
🚀 Enhancement
core
- Other
app-core, product-core
- #4337 Allow moving
tracks and views up/down/to top/to bottom, and better click and drag track
re-ordering (@cmdcolin)
core, product-core
core, web-core
- #4284 Allow getting
stack trace error dialog from session.notify errors
(@cmdcolin)
🐛 Bug Fix
- Other
- #4318 Use
node-fetch-native to fix warning from JBrowse CLI on node 21+
(@cmdcolin)
- #4319 Fix Hi-C
rendering for some high resolution files
(@cmdcolin)
- #4314 Fix loading of
BED12 data from a plaintext BED with column headers
(@cmdcolin)
- #4293 Fix alignment
curves showing up in inkscape for breakpoint svg
(@cmdcolin)
- #4287 Fix snap
package and also add deb package (@cmdcolin)
- #4277 Fix usage of
--assemblyNames in
jbrowse add-connection
(@cmdcolin)
- #4275 Fixes bug on
URL highlight param in which refName aliases were not working
(@carolinebridge-oicr)
text-indexing
Committers: 3