v2.2.1 Release
· 3 min read
Hello, we are happy to release v2.2.1!
This contains several improvements including
- Ability to display the reference sequence track as a "GC content" track
- Ability to view SyntenyTracks as a plain track on a regular linear genome view, and to create synteny views from this
It also contains two important bug fixes
- In v2.2.0 CRAM features did not display mismatches correctly on alignments tracks
- In v2.2.0 the LGV ImportForm froze when changing assemblies that had the same refnames
We recommend upgrading to fix these issues!
Here are some screenshots of the new features in this release as well
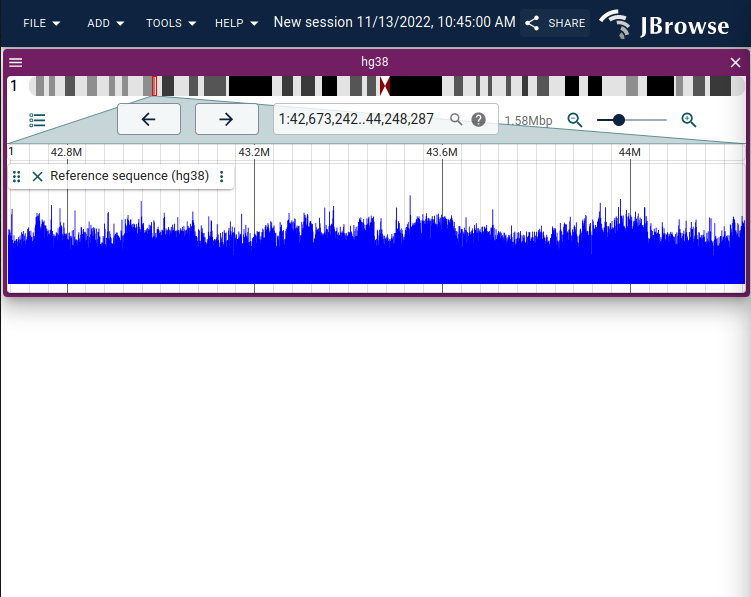
screenshot showing the GC content display on the reference sequence track
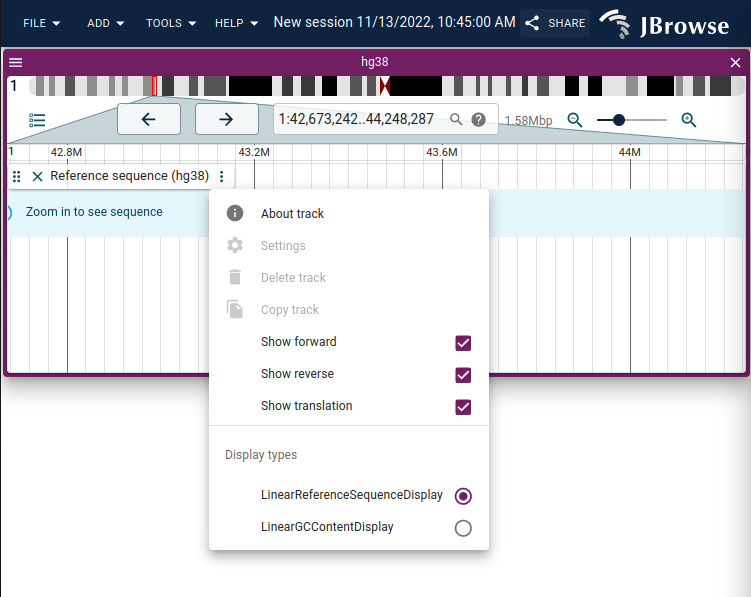
menu items for opening the GC content

screenshot showing right click on a feature in a SyntenyTrack -> Open synteny view

screenshot showing large synteny features on a SyntenyTrack in the LGV using a hg19 vs hg38 chain liftover file
Downloads
To install JBrowse 2 for the web, you can download the link above, or you can use the JBrowse CLI to automatically download the latest version. See the JBrowse web quick start for more details.
2.2.1 (2022-11-21)
🚀 Enhancement
- Other
core- #3298 Add authentication plugin to embedded components (@garrettjstevens)
- #3329 Add ability to minimize/collapse tracks and views and move views up/down in view stack (@cmdcolin)
- #3308 Add ability to display synteny track in a normal LGV showing regions of synteny as features (@cmdcolin)
- #3317 Move ErrorBoundary so that tracks/views that have crashed can be closed more easily (@cmdcolin)
🐛 Bug Fix
- Other
core
Committers: 2
- Colin Diesh (@cmdcolin)
- Garrett Stevens (@garrettjstevens)