v1.6.8 Release
· 3 min read
We are pleased to announce the v1.6.8 release!
This release includes a change to the default gene style to use boxes instead of "chevron" features, with a directional arrowhead
Before
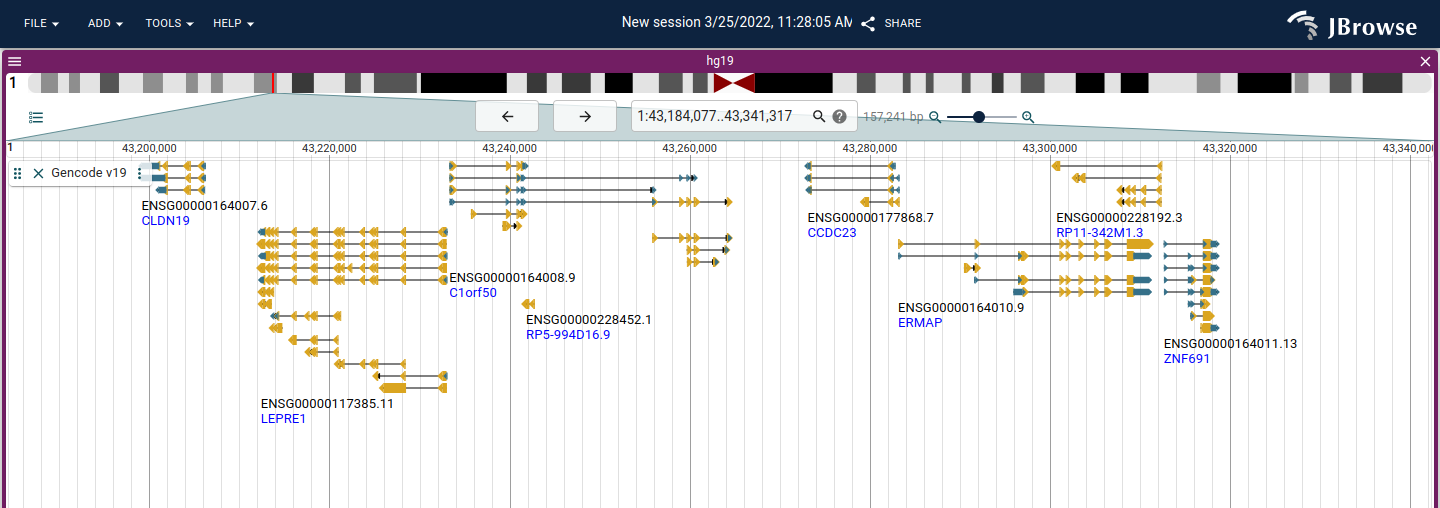
After

Some additional highlights
- Draws the correct the proportion of SNPs when displaying the SNPCoverage height in log scale (thanks @gringer for reporting)
- Adds more optimizations for some alignments tracks
- Adds a per-base drawing mode to alignments tracks

Downloads
To install JBrowse 2 for the web, you can download the link above, or you can use the JBrowse CLI to automatically download the latest version. See the JBrowse web quick start for more details.
1.6.8 (2022-03-25)
🚀 Enhancement
- Other
- #2847 Add option to color all the letters on all the reads to the pileup renderer (@cmdcolin)
- #2849 Avoid drawing intron subfeatures for gene glyphs (@cmdcolin)
- #2835 Hide add track and connection menu items when using embedded component (@carolinebridge-oicr)
- #2836 Display low-quality modifications in SNPCoverage renderer for MM tag (@cmdcolin)
core
🐛 Bug Fix
- Other
- #2852 Fix misaligned features under breakpoint split view (@cmdcolin)
- #2844 Fix layout of small features without labels for SvgFeatureRenderer (@cmdcolin)
- #2839 Fix the drawing of SNP height when the SNPCoverage track is using log scale (@cmdcolin)
- #2825 Fix tracklabels positioning not updating in UI after user selection (@cmdcolin)
core- #2841 Fix alignments tracks loading excessive data on chromosomes where no features exist (@cmdcolin)
- #2829 Allow user to specify number of workers (@garrettjstevens)
Committers: 3
- Caroline Bridge (@carolinebridge-oicr)
- Colin Diesh (@cmdcolin)
- Garrett Stevens (@garrettjstevens)